Keywords
personalised medicine, molecular diagnostics, genomics, cancer, clinical oncology, Ireland
Molecular diagnostics are critical for informing cancer patient care. In Ireland, the National Cancer Control Programme (NCCP) develops cancer therapy regimens, which include relevant information on molecular indications. Here, we present a collated overview of the current molecular indications of all NCCP systemic anti-cancer therapy regimens and the funding statuses of their associated drugs. Furthermore, we also provide estimates for the scale of required molecular testing in cancer therapy and for the clinical genetic sequencing capacity of Ireland, and provide a summary of current cancer clinical trials in Ireland which have molecular components.
Through a combination of web scraping, keyword search, and manual review, we performed a full review of all 856 indications included in the 533 therapy regimens published to date by the NCCP to identify therapy indications with explicit molecular criteria. For all cancer types identified in these indications, we obtained incidence rates in Ireland from National Cancer Registry Ireland to predict the number of patients yearly who stand to benefit from a molecular test. We then applied molecular subtype rates from published literature to estimate the number of patients who would then qualify for a relevant molecularly guided therapy.
We identified 246 indications for 175 NCCP therapy regimens that include molecular criteria. These 246 molecular indications encompassed 101 genetic criteria, 161 cellular biomarker criteria, 63 molecularly informed drugs, and over 20 cancer types. We estimated that up to approximately 55% of cancer patients in Ireland could qualify for a molecular test and that the majority of tested patients would qualify for a treatment informed by a molecular test.
As personalised cancer medicine continues to develop in Ireland, this study will provide a baseline understanding of current practices. We anticipate that work such as this will help to inform planning in the healthcare system.
personalised medicine, molecular diagnostics, genomics, cancer, clinical oncology, Ireland
In this revision, we have updated the included regimens and indications from November 2023 to May 2025. In addition, we have expanded on and clarified the testing criteria for some of these indications, as specified by the recently developed National Genomics Test Directory for Cancer.
See the authors' detailed response to the review by George Thomas
Modern genetics and genomics have played a vital role in human health for decades. However, since the advent of high-throughput next-generation sequencing (NGS), the role of genomics and molecular diagnostics in healthcare has increased dramatically1. As the science, engineering, and data analysis surrounding genomics continue to develop through research and innovation, genomics technologies progressively move from research and development into practical clinical usage in applications ranging from neonatal screening2 and hereditary disease risk3 to chemotherapy management and prognostics4.
To facilitate the integration of genomics and healthcare, many nations are in the process of developing or implementing strategies, legislation, policy, and infrastructure for clinical genomics5–9. Ireland is among these nations, having recently published a national plan for genomics medicine under the National Genomics and Genetics Strategy, which will oversee and guide implementation of the strategy as part of the national healthcare system in coming years10.
While science and innovation drive novel technologies and techniques in genomics, familiarity with current clinical practices is vital to matching research effort and expertise to clinical need and application. Here we aim to highlight actionable and informative molecular diagnostics in use in clinical oncology in Ireland by examining the cancer therapies and clinical trials currently informed by molecular diagnostics in Ireland. In addition, amidst increasing cancer incidence each year, we predict the number of patients in Ireland requiring a molecular diagnostic yearly and the number that would potentially benefit from molecular diagnostics and compare this to the availability of NGS infrastructure in major hospitals around the country.
Under the Health Service Executive (HSE), the National Cancer Control Programme (NCCP) is the leading national body addressing the diagnosis and treatment of cancer in Ireland. With the principal aim of implementing the Irish National Cancer Strategy, the NCCP's activities include reviewing new cancer therapies and developing national regimens for their use as part of the National Cancer Information System11.
New cancer drugs approved by the European Medicines Agency are assessed by the National Centre for Pharmacoeconomics, Ireland (NCPE) to produce a health technology assessment (HTA), which addresses the benefit vs. financial cost of the drug in question and recommends whether the drug should or should not be reimbursed by the HSE12. These reports, as well as information from experts and research, are assessed by the NCCP Technology Review Committee to recommend cancer drugs for funding under HSE drug schemes such as the Oncology Drugs Management Scheme (ODMS) or the Primary Care Reimbursement Services (PCRS) community drugs schemes13,14.
Independent of the funding status of a drug, the NCCP also develops, manages, and reviews national drug regimens addressing when and how these drugs should be used. In addition to information about drug combinations and dosing, these regimens also include, when relevant, the molecular indications required for the use of certain drugs in particular cancer types15. Note that while these regimens set guidelines for therapy, they are not exhaustive and clinical practice may differ when appropriate. As of July 2024, the NCCP and National Genetics and Genomics Office have also developed the NCCP Genomics Test Directory for Cancer, which catalogues and details genomic testing for a number of cancer types16.
Cancer is, by nature, a disease of genetic origins17. Though there are many different genome sequence mutations associated with many cancer types, only a small subset of these are currently known to be clinically informative or actionable, typically by informing diagnosis, prognosis, and/or treatment options18. For example, the EGFR gene encodes a tyrosine kinase which, when activated, signals for increased DNA replication and general cell proliferation; as such, over-activation of EGFR is associated with a variety of cancer types, including non-small cell lung cancer (NSCLC), in which approximately 14% of European patients harbour an EGFR-activating mutation19. For these patients, tyrosine-kinase inhibitor (TKI) therapies specifically targeting EGFR (e.g., osimertinib, gefitinib) are more effective and are associated with more favourable outcomes compared to chemotherapy20,21. In colorectal cancer patients, however, the presence of KRAS-activating mutations greatly reduces the efficacy of anti-EGFR TKI chemotherapies, as KRAS is a downstream activation target of the EGFR signalling pathway; once permanently activated through mutation, KRAS promotes tumour growth regardless of EGFR inhibition, and is associated with poorer outcomes22.
As in these examples, identifying genetic mutations can be critical in directing cancer treatment. Among all cancer treatment regimens developed by the NCCP, there are currently approximately 20 genetic factors (including 2 broader genetic phenotypes) informing 101 therapy indications across 72 chemotherapy regimens. These regimens involve combinations of 47 different genomics-informed drugs, 40 of which are approved for funding through either the PCRS or the ODMS for approved indications (Table 1)15,16,23–26.
Genetic indications per cancer type are listed with their associated drugs and drug reimbursement status in Ireland. (NSCLC: non-small cell lung cancer; mCRC: metastatic colorectal cancer; mCRPC: metastatic castration-resistant prostate cancer; ALL: acute lymphoblastic leukaemia; CLL: chronic lymphocytic leukaemia; AML: acute myeloid leukaemia; CML: chronic myelogenous leukaemia; GIST: gastrointestinal stromal tumour; PCRS: Primary Care Reimbursement Service; ODMS: Oncology Drugs Management System; MSI-H: microsatellite instability-high; dMMR: deficient mismatch repair; HRD: homologous recombination deficiency; ITD: internal tandem duplication) *Genomic instability includes genome-wide loss of heterozygosity, telomeric allelic imbalance, and large-scale transitions, as defined by the Myriad Genetics MyChoice CDx Plus assay27.
| Cancer Type | Subtype | Indication | Drugs | Reimbursement |
|---|---|---|---|---|
| Breast | Metastatic breast cancer | BRCA1/2 germline mutation | talazoparib | PCRS |
| High risk early breast cancer | BRCA1/2 germline mutation | olaparib | PCRS | |
| Lung | NSCLC | ALK fusion | alectinib | PCRS |
| brigatinib | PCRS | |||
| ceritinib | PCRS | |||
| crizotinib | PCRS | |||
| lorlatinib | PCRS | |||
| EGFR-activating mutation | afatinib | PCRS | ||
| dacomitinib | PCRS | |||
| erlotinib | PCRS | |||
| erlotinib and bevacizumab | erlotinib: PCRS; bevacizumab: Hospital | |||
| gefitinib | Hospital | |||
| osimertinib | PCRS | |||
| EGFR T790M mutation | osimertinib | PCRS | ||
| wild-type EGFR and ALK | atezolizumab | ODMS | ||
| ipilimumab and nivolumab | ODMS | |||
| pembrolizumab | ODMS | |||
| ROS1 mutation | crizotinib | Reimbursement for indication not approved | ||
| entrectinib | PCRS | |||
| MET exon 14 skipping mutation | tepotinib | PCRS | ||
| Gastrointestinal | mCRC | wild-type KRAS and NRAS | cetuximab | Hospital |
| panitumumab | Hospital | |||
| MSI-H or dMMR (including MLH1 promoter hypermethylation, PMS2, MSH2, MSH6) | ipilimumab and nivolumab | ODMS | ||
| pembrolizumab | ODMS | |||
| Cholangiocarcinoma | FGFR2 fusion or rearrangement | pemigatinib | Hospital | |
| Pancreatic adenocarcinoma | BRCA1/2 germline mutation | olaparib | PCRS | |
| Gynaecological | Epithelial ovarian, fallopian tube, and peritoneal cancers | HRD+ (BRCA1/2 somatic mutation or genomic instability*) | olaparib and bevacizumab | olaparib: PCRS; bevacizumab: Hospital |
| BRCA1/2 germline or somatic mutation | olaparib | PCRS | ||
| Endometrial cancer | MSI-H or dMMR (including MLH1 promoter hypermethylation, PMS2, MSH2, MSH6) | dostarlimab | Hospital | |
| Genitourinary | mCRPC | BRCA1/2 germline or somatic mutation | olaparib | PCRS |
| niraparib and abiraterone acetate (Akeega®) | PCRS | |||
| Metastatic urothelial carcinoma | FGFR3 mutation | erdafitinib | PCRS | |
| Sarcoma | GIST | CD117 mutation | imatinib | PCRS |
| Skin | metastatic melanoma | BRAF V600 mutation | dabrafenib | PCRS |
| dabrafenib and trametinib | PCRS | |||
| encorafenib and binimetinib | PCRS | |||
| vemurafenib | PCRS | |||
| vemurafenib and cobimetinib | PCRS | |||
| Any solid tumour | NTRK fusion | entrectinib | PCRS | |
| larotrectinib | PCRS | |||
| Leukaemia | ALL | BRC-ABL1 fusion | inotuzumab ozogamicin | ODMS |
| BRC-ABL1 fusion with T315I mutation | ponatinib | PCRS | ||
| BRC-ABL1 fusion negative | blinatumomab | ODMS | ||
| CLL | TP53 mutation or deletion | acalabrutinib | PCRS | |
| idelalisib and rituximab | idelalisib: PCRS; rituximab: Hospital | |||
| ibrutinib | PCRS | |||
| venetoclax | PCRS | |||
| zanubrutinib | PCRS | |||
| AML | FLT3 mutation | midostaurin | PCRS | |
| FLT3-ITD | quizartinib | PCRS | ||
| CML | BRC-ABL1 fusion | asciminib | PCRS | |
| bosutinib | PCRS | |||
| BRC-ABL1 fusion with T315I mutation | ponatinib | PCRS |
Depending on clinical purpose and cost, testing for relevant genetic mutations in cancer occurs at several levels of scale. Small-scale single-gene tests can be used to identify known point mutations, such as EGFR T790M or KRAS G12C chemotherapy resistance mutations28,29, or to identify known fusion genes, such as the BCR-ABL1 gene fusion found in chronic myelogenous leukaemia (CML)30. These single-gene tests are generally performed using techniques such as quantitative polymerase chain reaction (qPCR) or fluorescent in-situ hybridization (FISH), and can also be performed on both Sanger sequencing and next-generation sequencing (NGS) platforms, though using high-throughput NGS with very small targets is generally not cost efficient without very large numbers of samples.
Multiple genes can be tested for mutations simultaneously by sequencing on an NGS instrument. These NGS assays range from small disease-focused gene panels, targeting tens to hundreds of genes; to whole exome sequencing (WES or WXS), targeting tens of thousands of genes; to whole genome sequencing (WGS), which generates data from both genic and non-genic regions. In all NGS applications, results can then be subset virtually to focus on disease-specific genes or regions of interest. While methods like qPCR or genotyping microarrays can be used to detect known mutations, genome sequencing does not require a priori knowledge of mutations of interest, thus allowing for discovery of novel relevant genomic variation in cancer31. While novel mutations are not likely to be clinically actionable upon discovery, they may have potential for use in research, trials, and treatment in the future. More comprehensive genomic sequencing additionally allows for more complex genomic profiling strategies which can further inform disease aetiology, progression, and prognosis.
In Ireland, qPCR and FISH single-gene tests, gene panels including ThermoFisher's Oncomine Focus panels and other ThermoFisher Ion AmpliSeq small gene panels, and clinical exome gene panels are all routinely used. While whole genome sequencing can be clinically useful, this is generally not performed in Ireland as routine care outside of clinical trials or research applications.
In addition to the genetic sequencing performed by Irish medical laboratories, patient samples are also sent to external sequencing facilities in cases requiring, for example, rapid turnaround time, Sanger sequencing variant confirmation, or specialty assay sequencing. Notably, homologous recombination deficiency (HRD) has recently been added as an NCCP indication for olaparib treatment of ovarian cancer. While largely determined by the presence of deleterious BRCA1/2 mutations, HRD is a wider genetic phenotype influenced by larger genomic factors such as loss-of-heterozygosity and rearrangement events. Similarly, high microsatellite instability (MSI-H), which was recently added as an NCCP indication for immune checkpoint inhibitors in colorectal cancer, requires profiling of multiple locations throughout the genome. Both HRD and MSI-H testing thus require larger or specialty NGS gene panels, such as the Myriad MyChoice HRD test, FoundationOne panel, and Illumina TSO500 panel, all of which are currently being considered for use in Ireland. These external tests are generally funded under hospital departmental budgets rather than being reimbursed directly by the HSE, though efforts are underway by several hospitals to develop the infrastructure required to perform more genetic tests domestically in public facilities.
In addition to identifying mutated cancer-associated genes, confirming the presence of cellular biomarkers, which commonly include hormone receptors and antigens involved in immune cell recognition, can also be vital for accurate cancer diagnosis and treatment decisions. Lymphoma subtypes, for example, each exhibit characteristic immunophenotypes which can be essential for differential diagnosis of cancers that are otherwise morphologically similar32,33.
Biomarkers expressed on the cell surface can also serve as key drug targets. Antibody-based therapies target only specific cell types exhibiting the target antigen, and thus can activate or inhibit cellular signalling pathways or elicit a patient immune response against target cells, while limiting the potential deleterious effects of cancer treatment. Antibody-drug conjugates, such as brentuximab vedotin, further exploit this specificity by directing otherwise highly toxic chemotherapy drugs only to cells exhibiting the target antigen34.
Complementary to antibody-based therapies, small molecule drugs can also reach intracellular targets. For example, several treatment routes exist to reduce the growth-promoting effect of oestrogen on oestrogen-receptor-positive (ER+) breast tumours, including anastrozole, which binds aromatase enzymes to inhibit the production of oestrogen in the body; tamoxifen, which inhibits oestrogen binding by blocking oestrogen receptors; and fulvestrant, which binds and destabilises oestrogen receptors, inducing their breakdown by the cell35.
Like genetic mutations, the presence or absence of cellular biomarkers can play a critical role in diagnosis, prognosis, and treatment of a patient. In Ireland, the presence or absence of 10 markers are a factor for 161 indications for 25 different therapies across 117 treatment regimens published by the NCCP, and are of particular importance for informing breast cancer and lymphoma treatments, which account for 70% of these indications. Of the 25 included therapies, 21 have funding through the ODMS and PCRS (Table 2)15,16,23–26.
Indications are listed per cancer type with their associated drugs and drug reimbursement status in Ireland. (NSCLC: non-small cell lung cancer; mCRC: metastatic colorectal cancer; GEJ: gastro-oesophageal junction; HNSCC: head and neck squamous cell carcinoma; B-ALL: B-cell acute lymphoblastic leukaemia; AML: acute myeloid leukaemia; NHL: non-Hodgkin lymphoma; PCRS: Primary Care Reimbursement Service; ODMS: Oncology Drugs Management System; IHC: immunohistochemistry; ISH: in situ hybridisation; IC: immune cell score; TC: tumour cell score; TPS: tumour proportion score; CPS: combined positive score).
| Cancer Type | Subtype | Indication | Drugs | Reimbursement |
|---|---|---|---|---|
| Breast | ER+ | fulvestrant | PCRS | |
| tamoxifen | PCRS | |||
| HR+ | anastrozole | PCRS | ||
| exemestane | PCRS | |||
| letrozole | PCRS | |||
| HR+, HER2- | exemestane | PCRS | ||
| aromatase inhibitor or fulvestrant | PCRS | |||
| HER2+ (3+ by IHC, or ≥ 2 by ISH)46 | lapatinib | PCRS | ||
| neratinib | PCRS | |||
| trastuzumab | Hospital | |||
| trastuzumab and pertuzumab | trastuzumab: Hospital; pertuzumab: ODMS | |||
| trastuzumab deruxtecan | ODMS | |||
| trastuzumab emtansine (Kadcyla®) | ODMS | |||
| trastuzumab/pertuzumab (Phesgo®) | ODMS | |||
| HER2 low (1+ by IHC, or IHC2+/ISH negative)46 | trastuzumab deruxtecan | Reimbursement for indication not approved | ||
| HER2- | olaparib | PCRS | ||
| PD-L1+ (SP142 Ventana: ≥ 1% IC), HER2-, HR- | atezolizumab | ODMS | ||
| Lung | NSCLC | PD-L1+ (SP142 Ventana: ≥ 50% TC or ≥ 10% IC), HER2-, HR- | atezolizumab | ODMS |
| PD-L1+ (SP263 Ventana: ≥ 1% TC) | durvalumab | ODMS | ||
| PD-L1+ (SP263 Ventana: ≥ 1% TC) | nivolumab | ODMS | ||
| PD-L1+ (SP263 Ventana: ≥ 50% TPS) | pembrolizumab | ODMS | ||
| Gastrointestinal | mCRC | EGFR+ | cetuximab | Hospital |
| Metastatic stomach adenocarcinoma | HER2+ | trastuzumab | Hospital | |
| Metastatic gastric or GEJ cancer | HER2+ | trastuzumab | Hospital | |
| Metastatic gastric cancer, GEJ cancer, or oesophageal adenocarcinoma | PD-L1+ (28-8 Dako: CPS ≥ 5), HER2- | nivolumab | Reimbursement for indication not approved | |
| Oesophageal squamous cell carcinoma | PD-L1+ (28-8 Dako: ≥ 1% TC) | nivolumab | ODMS | |
| GEJ adenocarcinoma | PD-L1+ (22C3 Dako: CPS ≥ 10), HER2- | pembrolizumab | ODMS | |
| Oesophageal carcinoma | PD-L1+ (22C3 Dako: CPS ≥ 10) | pembrolizumab | ODMS | |
| Genitourinary | Urothelial carcinoma | PD-L1+ (SP142 Ventana: ≥ 5% IC) | atezolizumab | ODMS |
| PD-L1+ (28-8 Dako: ≥ 1% TC) | nivolumab | ODMS | ||
| PD-L1+ (22C3 Dako: CPS ≥ 10) | pembrolizumab | ODMS | ||
| Gynaecological | Cervical cancer | PD-L1+ (CPS ≥ 1) | pembrolizumab | Reimbursement by exception |
| Head & Neck | HNSCC | PD-L1+ (CPS ≥ 1) | pembrolizumab | ODMS |
| Leukaemia | B-ALL | CD19+ | blinatumomab | ODMS |
| CD22+ | inotuzumab ozogamicin | ODMS | ||
| AML | CD33+ | gemtuzumab ozogamicin | ODMS | |
| Lymphoma | Hodgkin lymphoma | CD30+ | brentuximab vedotin | ODMS |
| CD20+ | rituximab | Hospital | ||
| Non-Hodgkin B-cell lymphomas | CD20+ | rituximab | Hospital | |
| Follicular lymphoma | CD20+ | obinutuzumab | ODMS | |
| CD20+ | rituximab | Hospital | ||
| Systemic anaplastic large cell lymphoma | CD30+ | brentuximab vedotin | ODMS | |
| Cutaneous T-cell lymphoma | CD30+ | brentuximab vedotin | ODMS |
The presence of cellular biomarkers can be determined either by direct detection, or by some indirect indication of their presence. Immunohistochemistry (IHC) techniques, which remain the gold standard for direct determination, use a combination of an antigen-specific antibody and a dye or fluorophore to indicate antigen presence in cancer tissue samples via microscopy36. While this technique can generally only detect one antigen per assay, the process can be parallelized in appropriate tissue samples via flow cytometry, such that multiple antibodies can be applied, allowing several antigens to be detected on cancer cells simultaneously37,38.
Indirect detection, instead, can be accomplished through gene expression analysis. Rather than detecting an antigen of interest via an antibody, this approach involves the quantification of RNA transcripts encoding the biomarker. Techniques for measuring expression analysis are similar to those for detecting DNA mutations and include reverse transcription qPCR (RT-qPCR), expression microarrays, and next-generation RNA sequencing (RNA-seq).
These methods also scale similarly to DNA mutation detection methods: qPCR is limited to measuring the expression of single genes, while microarrays and RNA-seq are able to simultaneously quantify thousands of transcripts. Of particular note is that RNA-seq, in addition to expression analysis, also allows for mutation detection by default. This includes more complex mutations such as fusion genes, which are frequently highly associated with cancer and can serve as drug targets for inhibitors such as ponatinib, which inhibits BCR-ABL1 fusion proteins found in CML39, and larotrectinib, a novel tumour-agnostic Trk inhibitor that can be used for any cancer in which NTRK-family fusions are detected40. While RNA expression assays are less commonly used in clinical practice, recent studies have shown comparable test results between RNA-seq and IHC41.
In Irish hospitals, cellular biomarker detection methods generally include single gene tests like IHC and RT-qPCR. In addition, the external testing service Oncotype DX® is available in Ireland for breast cancer patients and uses RT-qPCR to measure the expression of 16 genes, including HER2 and both oestrogen and progesterone receptor genes42–44. While RNA-seq remains uncommon, reimbursement for larotrectinib in Ireland notably requires submission of RNA-seq results45.
Data on the incidence of cancer in Ireland has been centrally recorded by National Cancer Registry Ireland (NCRI) since 1994. While the total incidence of cancer in Ireland has doubled since 1994 (Figure 1a, 1c), the rate of cancer incidence has increased by approximately 50% (Figure 1b) and age-adjusted incidence has increased by approximately 15% (Figure 1d), reflecting at least in part the advancing age profile of the larger population and increases in life expectancy47,48. Latest available figures (from 2020) show a current 1 in 2 lifetime risk of invasive cancer49. Fortunately, overall cancer survival in Ireland has also increased (Figure 2), with a gain of approximately 15 percentage point survivorship over the same time period50.
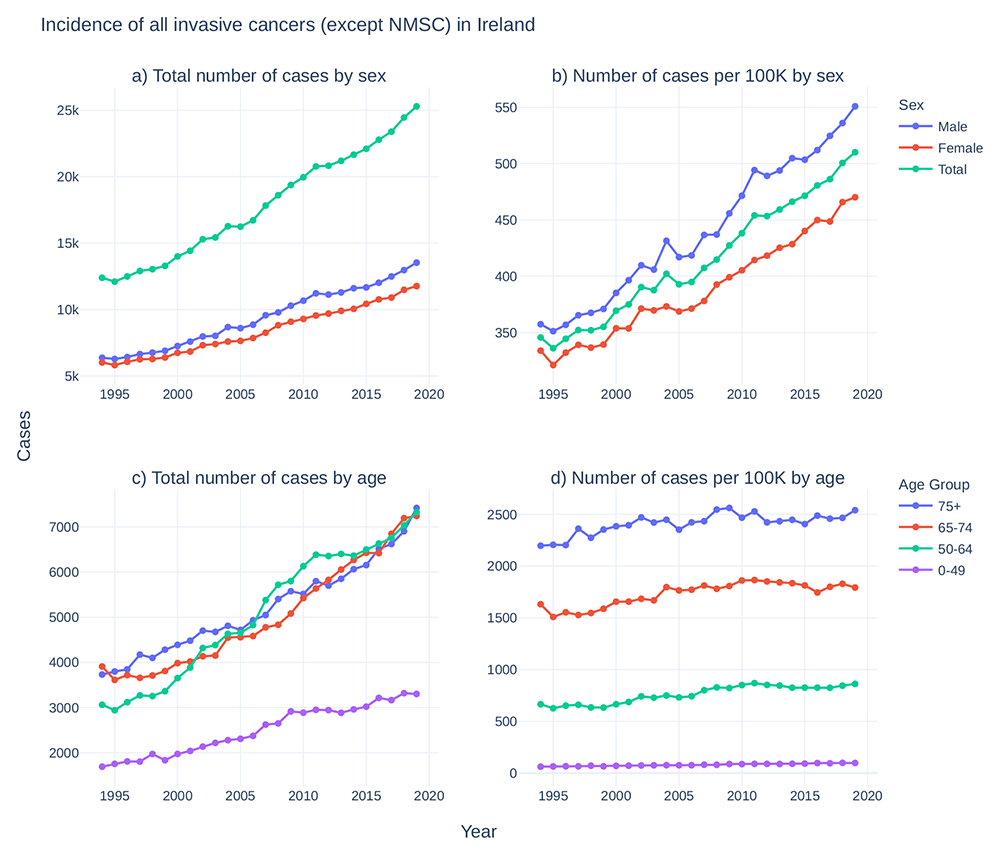
Top: For males (blue), females (red), and total population (green), a) case counts of cancers per year and b) case counts per 100,000 individuals in each category per year. Bottom: For 0–49 (purple), 50–64 (green), 65–74 (red), and 75+ (blue) years old, c) cancer case counts per year and d) cancer case counts per 100,000 individuals in each category per year. Cancer incidence data taken from National Cancer Registry Ireland47. National population estimates per year taken from the Central Statistics Office Ireland48. (NMSC: non-melanoma skin cancer).
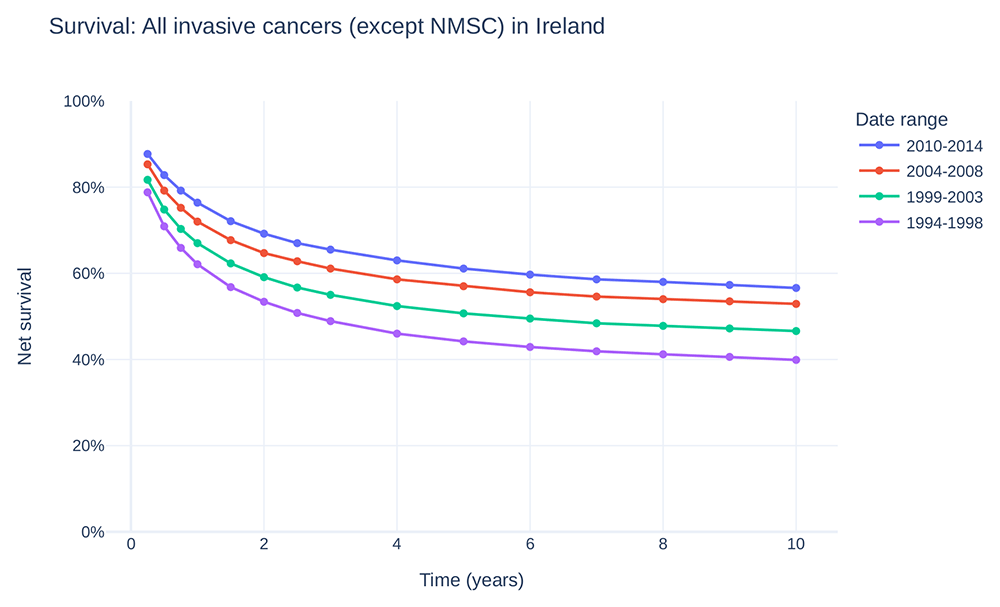
Survival curve estimates showing percent net survival at selected time points for diagnoses made during the given time period. Data provided by and plot adapted from National Cancer Registry Ireland47. (NMSC: non-melanoma skin cancer).
Advances in personalised medicine continue to contribute to this survival improvement, with cancer molecular diagnostics enabling a wide range of modern therapy options. However, it is not clear how many patients in Ireland currently receive or stand to benefit from molecular cancer diagnostics. Data on the rate of molecular diagnostics usage in Ireland is not publicly available and is not currently centrally recorded. This information is relevant to quantification of the potential benefits of genomic tests on the population level and for resource planning, not least as part of the National Genomics and Genetics Strategy. Furthermore, while NCRI collects and provides information on cancer incidence, specific cancer rates are categorised by International Classification of Diseases 10th revision (ICD-10) definitions largely classed by tissue type, leaving molecular subtype rates in Ireland unknown.
Based on current disease-informing molecular diagnostics listed in NCCP treatment regimens, cancer incidence rates published by NCRI, published studies on molecular cancer subtypes, and the single most common molecular subtype per cancer, we estimate that over 13,000 patients should be receiving some form of molecular diagnostic test yearly to identify the subpopulation of at least 7,000 cancer patients that stand to benefit from current molecular-diagnostic-guided therapeutics used in Ireland. These include over 2,000 patients who would qualify for a genetic-guided therapy, and 6,500 patients who would qualify for a cellular biomarker-guided therapy (Table 3 and Table 4). This testing burden represents approximately 55% of the 24,000 invasive cancer cases in Ireland yearly (excluding non-melanoma skin cancer), with about 30% directly benefiting from a test result49. It should be noted that these numbers only include tests that directly inform therapy, and do not include the large body of molecular tests performed primarily for diagnostic or prognostic purposes. In addition, NTRK-fusion testing is not included in these estimates, as treatment is tumour-agnostic and NTRK-fusion incidence is quite variable across various tumour types51.
Yearly incidence of cancer types with a genetic indication in Ireland, with predicted numbers of positive molecular diagnoses based on rates from literature. Incidence in Ireland published by National Cancer Registry Ireland49, unless otherwise noted by citation. Molecular subtype rate estimates (MD+ Rate) for each cancer obtained from indicated references. (ICD-10: International Classification of Diseases 10th Revision; MD: Molecular diagnostic; NSCLC: non-small cell lung carcinoma; mCRC: metastatic colorectal cancer; B-ALL: B-cell acute lymphoblastic leukaemia; CLL: chronic lymphocytic leukaemia; AML: acute myeloid leukaemia; CML: chronic myelogenous leukaemia; mCRPC: metastatic castration-resistant prostate cancer; GIST: gastrointestinal stromal tumour; MSI-H: microsatellite instability-high; dMMR: deficient mismatch repair; ITD: internal tandem duplication) *Only female cases included. **Chemotherapy resistance mutation incidence is variable, with incidence typically increasing in response to therapy.
| Cancer Type | ICD-10 Label | Subtype | Incidence in Ireland | Molecular Diagnostic (MD) | MD+ Rate | MD+ Incidence in Ireland | References |
|---|---|---|---|---|---|---|---|
| Breast | C50: Malignant neoplasm of breast | 3363* | BRCA1/2 germline mutation | 0.024 | 81 | 52–55 | |
| Lung | C34: Malignant neoplasm of bronchus and lung | NSCLC56–58 | 2268 | ALK fusion | 0.05 | 113 | 59–61 |
| METex14 skipping | 0.02 | 45 | 62 | ||||
| ROS1 mutation | 0.02 | 45 | 63 | ||||
| EGFR-activating mutation | 0.14 | 320 | 19,59 | ||||
| EGFR T790M mutation | Builds** | 64,65 | |||||
| Gastrointestinal | C18-21: Malignant neoplasm of: - colon - rectosigmoid junction - rectum - anus and anal canal | mCRC66 | 1058 | wild-type KRAS and NRAS | 0.39 | 416 | 66 |
| MSI-H or dMMR | 0.04 | 42 | 67–70 | ||||
| C25: Malignant neoplasm of pancreas | Pancreatic adenocarcinoma | 624 | BRCA1/2 germline mutation | 0.04 | 25 | 71 | |
| C22: Malignant neoplasm of liver and intrahepatic bile ducts | Cholangiocarcinoma72,73 | 52 | FGFR2 fusion or rearrangement | 0.14 | 7 | 74–77 | |
| Skin | C43: Malignant melanoma of skin | Melanoma | 1170 | BRAF V600 mutation | 0.5 | 585 | 78–80 |
| Genitourinary | C61: Malignant neoplasm of prostate | mCRPC81,82 | 534 | BRCA1/2 germline or somatic mutation | 0.14 | 75 | 83 |
| C65-68: Malignant neoplasm of: - renal pelvis - ureter - bladder - other and unspecified urinary organs | Urothelial carcinoma84–86 | 536 | FGFR3 mutation | 0.25 | 134 | 87,88 | |
| Gynaecological | C54: Malignant neoplasm of corpus uteri | Endometrial cancer | 538 | MSI-H or dMMR | 0.25 | 135 | 89,90 |
| C56: Malignant neoplasm of ovary | Epithelial ovarian cancer91–93 | 361 | BRCA1/2 germline or somatic mutation | 0.25 | 90 | 94–96 | |
| C57.0: Malignant neoplasm: Fallopian tube | Fallopian tube cancer | 25 | 0.35 | 9 | 94–96 | ||
| C48: Malignant neoplasm of retroperitoneum and peritoneum | Peritoneal carcinoma | 24* | 0.16 | 4 | 94–96 | ||
| Leukaemia | C91.0: Acute lymphoblastic leukaemia [ALL] | B-ALL97 | 49 | BCR-ABL1 fusion | 0.04 paediatric, 0.25 adult | 5 | 98 |
| BCR-ABL1 fusion with T315I mutation | Builds** | 99 | |||||
| C91.1: Chronic lymphocytic leukaemia of B-cell type | CLL | 202 | TP53 mutation or deletion | 0.1 | 20 | 100 | |
| C92.0: Acute myeloblastic leukaemia [AML] | AML | 138 | FLT3 mutation | 0.3 | 41 | 101,102 | |
| FLT3-ITD | 0.2 | 28 | 102,103 | ||||
| C92.1: Chronic myeloid leukaemia [CML], BCR/ABL-positive | CML | 63 | BCR-ABL1 fusion | 0.94 | 59 | 104 | |
| BCR-ABL1 fusion with T315I mutation | Builds** | 104 | |||||
| Sarcoma | C49: Malignant neoplasm of other connective and soft tissue | GIST105 | 20 | CD117 mutation | 0.8 | 16 | 106,107 |
| Total, max per cancer type | 11024 | 2022 |
Yearly incidence of cancer types with a cellular biomarker-based diagnostic in Ireland, with predicted numbers of positive molecular diagnoses based on rates from literature. Incidence in Ireland published by National Cancer Registry Ireland49, unless otherwise noted by citation. Molecular subtype rate estimates (MD+ Rate) for each cancer obtained from indicated references. (ICD-10: International Classification of Diseases 10th Revision; MD: Molecular diagnostic; NSCLC: non-small cell lung carcinoma; mCRC: metastatic colorectal cancer; GEJ: gastro-oesophageal junction; ALCL: anaplastic large cell lymphoma; CTCL: cutaneous T-cell lymphoma; HNSCC: head and neck squamous cell carcinoma; B-ALL: B-cell acute lymphoblastic lymphoma; AML: acute myeloid leukaemia) *Only female cases included.
| Cancer Type | ICD-10 Label | Subtype | Incidence in Ireland | Molecular Diagnostic (MD) | MD+ Rate | MD+ Incidence in Ireland | References |
|---|---|---|---|---|---|---|---|
| Breast | C50: Malignant neoplasm of breast | 3363* | HR+ | 0.818 | 2751 | 108 | |
| ER+ | 0.806 | 2711 | 108 | ||||
| HER2+ | 0.154 | 517 | 108 | ||||
| HER2-low | 0.4 | 1345 | 109,110 | ||||
| PD-L1+ | 0.197 | 663 | 111 | ||||
| Lung | C34: Malignant neoplasm of bronchus and lung | NSCLC56–58 | 2268 | PD-L1+ | 0.326 | 739 | 112,113 |
| Gastro-intestinal | C15: Malignant neoplasm of oesophagus C16: Malignant neoplasm of stomach | Metastatic gastric cancer, GEJ cancer, or oesophageal adenocarcinoma | 1072 | PD-L1+, HER2- | 0.45 | 482 | 114 |
| C16: Malignant neoplasm of stomach | Stomach or GEJ cancer | 557 | HER2+ | 0.221 | 123 | 115 | |
| C15: Malignant neoplasm of oesophagus | Oesophageal or GEJ cancer | 515 | PD-L1+ | 0.45 | 232 | 116–119 | |
| C18-21: Malignant neoplasm of: - colon - rectosigmoid junction - rectum - anus and anal canal | mCRC66 | 1058 | EGFR+ | 0.6 | 635 | 120,121 | |
| Lymphoma | C82: Follicular lymphoma C83: Non-follicular lymphoma C85: Other and unspecified types of non-Hodgkin lymphoma C88: Malignant immunoproliferative diseases | Non-Hodgkin B-cell lymphomas | 712 | CD20+ | 0.98 | 698 | 122 |
| C81: Hodgkin lymphoma | Hodgkin lymphoma123 | 127 | CD30+ | 1 | 127 | 32,123 | |
| C84: Mature T/NK-cell lymphomas | ALCL and CTCL | 83 | CD30+ | 1 | 83 | 124 | |
| Head & Neck | C00-14: Malignant neoplasms of lip, oral cavity and pharynx C30-C32: Malignant neoplasm of: - nasal cavity and middle ear - accessory sinuses - larynx | HNSCC | 786 | PD-L1+ | 0.85 | 668 | 125 |
| Genito- urinary | C65-68: Malignant neoplasm of: - renal pelvis - ureter - bladder - other and unspecified urinary organs | Urothelial carcinoma84–86 | 536 | PD-L1+ | 0.303 | 162 | 126,127 |
| Gynaecological | C53: Malignant neoplasm of cervix uteri | Cervical cancer | 253 | PD-L1+ | 0.85 | 215 | 128,129 |
| Leukaemia | C92.0: Acute myeloblastic leukaemia [AML] | AML | 138 | CD33+ | 0.85 | 117 | 130 |
| C91.0: Acute lymphoblastic leukaemia [ALL] | B-ALL97 | 49 | CD19+ | 1 | 49 | 131 | |
| CD22+ | 0.98 | 48 | 132 | ||||
| Total, max per cancer type | 10444 | 6849 |
To accommodate the clinical needs of these individuals, clinical laboratories in the Republic of Ireland operate several makes of instruments, each with their own capacity and throughput. In total, there are four Illumina NextSeq, one Illumina MiniSeq, and four ThermoFisher Ion Torrent NGS instruments currently operating in clinical practice across 5 Irish hospitals. In addition, there are a number of qPCR machines available for single-gene tests, as well as one Sanger sequencing platform for confirmation testing. Based on published technical specifications133,134, the combination of high-throughput NextSeq and Ion Torrent instruments in Ireland represent a maximum nominal capacity of approximately 44 – 68 deep whole exomes sequenced in a 30 hour period, depending on targeted depth, exome size, and amplification, though in reality this number is also greatly dependent upon sample batching, laboratory operation and sample preparation time, specific instrument configuration, and operating costs, among numerous other factors.
In addition to routine care pathways, clinical trials offer some patients access to cancer therapies that would not otherwise be available, typically because the therapy is novel or is not yet offered in Ireland. Clinical trials for cancer therapies dictate strict enrolment criteria, and these are often based on molecular diagnosis of cancer subtypes. In November 2023, Cancer Trials Ireland, for example, listed 86 clinical trials for cancer available in the country. Of these, at least 43 listed a molecular diagnostic as either eligibility criteria or as a factor in the trial (Table 5)135. For example, the KRYSTAL-10 and LOXO 101 trials were both active in Ireland: KRYSTAL-10 recruited at several Irish hospitals and investigated the use of a novel KRAS-inhibiting drug, adagrasib, to treat colorectal cancer patients who have the KRAS-activating G12C mutation136, while LOXO 101 investigated the use of larotrectinib to treat any cancer harbouring an NTRK gene fusion that has been confirmed via molecular assay137. Besides drug trials, other efforts in the field of genomics are also underway in clinical trials in Ireland, including fundamental research into the genetic profiling of cancers and DNA biobanking138.
Summary of cancer clinical trials listed by Cancer Trials Ireland in November 2023 whose study designs include a molecular component. Clinical trial IDs are given as clinicaltrials.gov IDs where available (except trial ITCC-059, which is listed by EudraCT ID). (miRNA: microRNA; GEJ: gastro-oesophageal junction; MIBC: muscle invasive bladder cancer; ctDNA: circulating tumour DNA; dMMR: deficient mismatch-repair; HNSCC: head and neck squamous cell carcinoma; AML: acute myeloid leukaemia; MDS-EB2: myelodysplastic syndromes with excess blasts-2; NSCLC: non-small cell lung carcinoma; NGS: next-generation sequencing; DLBCL: diffuse large B-cell lymphoma; CML: chronic myelogenous leukaemia; B-ALL: B-cell acute lymphoblastic leukaemia; CNS: central nervous system; ALL: acute lymphoblastic leukaemia; MDS: myelodysplastic syndrome; JNML: juvenile myelomonocytic leukaemia; HRRm: homologous recombination repair mutation; HRD: homologous recombination deficiency).
Molecular diagnostics, in the form of both genetic and cellular biomarker testing, are a vital component of cancer diagnostics and treatment. In Ireland, the NCCP lists 175 treatment regimens with a molecular diagnostic component, through which 30% of the Irish cancer patient population stands to directly benefit. Cancer cases are predicted to double in Ireland by 2045139, underscoring the need to ensure that the increasing requirement for testing is met by Irish infrastructure. As research highlights further drug repurposing and new off-label drug uses, as novel precision medicine therapies are produced against innovative drug targets in more cancer types, and as clinical trials become more widely available in Ireland, the need for molecular testing is likely to increase steadily until the total number of required molecular tests converges with, and exceeds, the total number of cancer cases. It should also be noted that these numbers do not include testing for inherited cancer risk or any non-cancer disease, each of which will add to the requirement for molecular diagnostics. While this presents a challenge to any national healthcare system, it promises great improvements in personalised cancer care and outcomes for patients in the near future if the challenge can be met.
Ireland's recent National Genomics and Genetics Strategy will represent the first major strides in addressing this challenge. While the strategy encompasses many aspects, a key consideration that should be highlighted is the need for a collaborative approach from all stakeholders. Fundamental to this approach must be the facilitation of a modernised, centralised exchange of expertise and data from all parties, including the NCCP and NCRI for cancer expertise and statistics, the NCPE for pharmacoeconomics, hospitals for current infrastructure and implementation, and universities for current research efforts.
For this strategy to be successful, decisions must be based on accurate data gathered by these institutions. While genomics initiatives and strategies in countries with comparable population sizes (such as the Precision Medicine Centre of Excellence in Northern Ireland140, the regional laboratories established through the Scottish Strategic Network for Genomic Medicine141, the hub-and-spoke model employed in Denmark7, or the distributed specialisation across institutions in Norway's InPreD initiative142) can inform Irish efforts, it is critical to collect and analyse healthcare data in Ireland to establish a viable and appropriate molecular medicine service capable of meeting Irish clinical demand. This data will be foundational for evaluating the utility of clinical care in Ireland moving forward, particularly in pharmacoeconomic areas such as health technology assessments, pharmaceutical pricing, and drug reimbursement approvals. Furthermore, national infrastructure to support the collection and storage of molecular patient data will enable Ireland to participate in international research initiatives, such as the European Commission's Digital Europe Call for genomics data, and the proposed EU European Health Data Space143.
In this article, we sought to collate available data from various sources across Ireland to present a unified overview of the state of cancer molecular diagnostics in Ireland. Ultimately, to best address Ireland's future need for molecular and genomic medicine, we first need to accurately establish Ireland's current capabilities and position, and it is our hope that others will follow in contributing to this effort.
NCCP cancer therapy regimens were accessed via the HSE NCCP National SACT Regimens website15. Information on therapy indications from each tumour group subpage (as well subpages for oral anti-cancer medicines and paediatric therapies) was collected by systematic HTML parsing of tabular elements using the Python package Beautiful Soup version 4.11.2 in Python 3.11.0144. Raw therapy indication text was then further parsed in Python to harmonise descriptions and drug names, to combine duplicate indications by indication ID, to assign relevant disease based on website subpage and subheadings, and to group therapy indications by regimen ID. Where conflicts arose in merging duplicate indications by ID, manual harmonisation was performed by referring to the full text of the hyperlinked regimen document; where conflicts arose in the hyperlinked regimen documents, the latest revision was used as reference. After tabular export of all indications and associated information, final manual curation was performed to correct malformed entries and errors in the source material, again referring to the appropriate full-text regimen documents. For the Python parser tool created for this purpose, see Software Availability145 and for the exported and manually curated data table, see also Software Availability23.
Identification of indications informed by genetic diagnostics was performed through several rounds of key-word search and manual review through the short descriptions of each therapy indication. Key-words included terms associated with genetics and genomics such as gene, chromosome, and express; known cancer gene names; and the terms and symbols positive, negative, +, and -, as well as further keywords encountered during manual review. In ambiguous cases, including cases where a molecular diagnostic was listed for one indication of a regimen, but not for other similar indications for the same regimen, both the full-text regimen document as well as published literature on the therapy in question were consulted. Note that while many regimens include CD20 antibody therapies for lymphoma, these were only included when a molecular diagnostic was explicitly referenced.
Reimbursement information was obtained from NCCP indications and regimens15, the NCCP table of approved drugs24, the PCRS list of reimbursable items25, and the HSE list of the High Tech Drug Arrangements26.
The regimen information in this article reflects the NCCP SACT Regimens website as of 2025-May-27.
Cancer incidence rates in Ireland were obtained from the NCRI publication Cancer in Ireland 1994–2020: Annual Statistical Report of the National Cancer Registry, Appendix I: Incident Cancer Cases49, except where indicated. Case numbers in this publication are listed as the 3-year average incidence from 2018–2020 of each ICD-10 invasive cancer group.
For each unique molecular diagnostic for each cancer subtype, incidence of the cancer subtype relative to the broader cancer type (e.g., proportion of lung cancers that are NSCLC) was obtained from literature where appropriate. Incidence rates of each molecular diagnostic within the relevant cancer subtype (e.g., proportion of NSCLC that is ALK+) were then also obtained from literature (references provided in Table 3 and Table 4). These rates were then applied to incident cancer rates in Ireland to estimate the positivity rate of each molecular diagnostic in Ireland.
In the case of acute lymphoblastic leukaemia, B-ALL subtype incidence was estimated separately for paediatric and adult cases due to differences in B- vs T-ALL rates in adults and children and the high proportion of childhood cases97. Similarly, separate molecular subtype rates were applied for BCR-ABL1 fusions in adult and childhood B-ALL for the same reason98. Incidence of metastatic castration-resistant prostate cancer was calculated as a function of total population based on the model referenced, producing numbers in agreement with NCRI case counts81,82. Rates of urothelial carcinoma were applied separately for primary urethral urothelial carcinoma due to lower published rates of urothelial histology84–86.
Technical specifications on the machine runtime and DNA throughput for the Illumina NextSeq and ThermoFisher Ion Torrent platforms were obtained from their respective manufacturer websites. To calculate nominal maximum throughput, the highest throughput configuration of each machine was used (NextSeq 550 High-Output = 100–120 Gb of 120 bp paired-end reads/29 hrs133, Ion GeneStudio with Ion 550 Chip = 40–50 Gb of 200 bp paired-end reads per 12 hrs134).
DNA sequencing target size was based on paired-end sequencing with a 120x coverage target using the Agilent SureSelect Clinical Research Exome V4 (total design size=51.0 Mb), for a targeted total of 12.24 Gb of genetic material sequenced per sample146.
Maximum capacity was then calculated to be the total number of exomes able to be sequenced by all 8 machines running at maximum capacity. The lower end of this range represents one run of each instrument using the lower bound of the instruments' stated throughput (one 29-hour run of the NextSeq at 100 Gb = 8 exomes per machine = 32 exomes, plus one 12-hour run of the Ion Torrent at 40 Gb = 3 exomes per machine = 12 exomes, totalling 44 exomes), while the higher end of the range represents the higher bound of the instruments' stated throughput, with two 12-hour runs of the Ion Torrent within the same time frame as one 29-hour NextSeq run (one 29-hour run of the NextSeq at 120 Gb = 9 exomes per machine = 36 exomes, plus two 12-hour runs of the Ion Torrent at 50 Gb = 8 exomes twice per machine = 32 exomes, totalling 68 exomes).
Information on clinical trials was obtained from Cancer Trials Ireland in November 2023135. Trials were considered to have a molecular component if the trial eligibility criteria included genetic mutations, aberrant genetic pathways, gene expression, cellular biomarkers, or microsatellite instability status as inclusion or exclusion criteria, or if the trial's purpose was to otherwise collect or analyse genomic data. Trials were evaluated systematically, beginning by prioritising those with explicit mention of these criteria in their short description. Trials without explicit reference to one of the two criteria, but which referenced a disease or treatment known to have a strong or common molecular diagnostic component were also prioritised. Short-listed trials' full trial descriptions were then checked to confirm the nature of the trial. After confirmation of short-listed trials, remaining trial full descriptions were then checked to confirm absence of a molecular diagnostic component.
Zenodo: Table of Indications and Regimens from the National Cancer Control Programme, Ireland. https://zenodo.org/doi/10.5281/zenodo.1015793923
This project contains the following underlying data:
Zenodo: NCCP-SACT-Parser https://zenodo.org/records/10660553
Python code to parse Ireland's National Cancer Control Programme cancer therapy indications for tabulation
This project contains the following extended data:
Data are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
Analysis code available from: https://github.com/TDMedina/NCCP-SACT-Parser145
Archived analysis code at time of publication: https://doi.org/10.5281/zenodo.10660553
License: Creative Commons Zero v1.0 Universal (CCO 1.0 Public domain dedication)
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Precision Oncology;Molecular Diagnostics; Cancer Therapeutics: Companion Diagnostics; Pathology
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
No source data required
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Precision Oncology;Molecular Diagnostics; Cancer Therapeutics: Companion Diagnostics; Pathology
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: molecular genetics, molecular diagnostics, non.invasive diagnostics, cell-free nucleic acids, liquid biopsy, cancer
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 09 Jun 25 |
read | |
|
Version 1 26 Mar 24 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Register with HRB Open Research
Already registered? Sign in
Submission to HRB Open Research is open to all HRB grantholders or people working on a HRB-funded/co-funded grant on or since 1 January 2017. Sign up for information about developments, publishing and publications from HRB Open Research.
We'll keep you updated on any major new updates to HRB Open Research
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)