Keywords
Peptide polymers; antimicrobial; diabetic foot infection; wound infection; biomaterials
Peptide polymers; antimicrobial; diabetic foot infection; wound infection; biomaterials
Chronic wound infections are common in community and healthcare settings and result in significant hospitalizations, morbidity and mortality. These infections include infected pressure sores, surgical site infections, burns and diabetic foot infections (DFI). DFIs are among the strongest predictors of lower limb amputation1 and annually, more than one million people with diabetes suffer lower limb loss following failure of therapeutic interventions for DFI2. Systemic antibiotic therapy is central to the range of multidisciplinary interventions for DFI. Despite the potential advantages of topical antimicrobial agents, such as targeting high concentrations directly to the site of infection and avoidance of toxic systemic effects, to date they have not been considered an effective replacement for systemic agents. However, there is a paucity of well-designed randomised controlled trials in this area3,4.
The microflora of chronic wounds is heterogenous and poly-microbial but the opportunistic pathogens, Staphylococcus aureus and Pseudomonas aeruginosa are among the most frequently isolated bacteria from these infections in humans5,6. In the setting of chronic wounds, these organisms produce biofilms, assembled communities of surface-bound cells enclosed in an extracellular matrix. In addition to resistance to phagocytosis, biofilms are recalcitrant to many antibiotics. This feature can complicate and significantly extend antibiotic treatment and may require higher antibiotic concentrations to reach the site of infection due to poor penetration. Peripheral arterial disease, if present, may be a further barrier to achieving biofilm-active concentrations7. Among S. aureus, the involvement of methicillin resistant S. aureus (MRSA) in these infections, with reported rates of up to 46 % in the USA8, further limits therapeutic options and often results in longer clinical courses and poorer outcomes. P. aeruginosa is resistant to multiple antibiotic classes, both inherently and through acquisition of antibiotic resistance genes. Given the combined challenges of multidrug resistance and biofilm involvement in chronic wound infections, therapeutic agents with mechanisms unrelated to those of conventional antibiotics that can penetrate and kill biofilms are urgently needed.
Structurally nano-engineered antimicrobial peptide polymers (SNAPPs) have received much attention as novel antimicrobials due to their potent antimicrobial activity which targets the cell membrane, unlike most conventional antibiotics. SNAPPs based on repeating units of valine and lysine arranged in a star-shape around a central core developed by Lam et al.9 have antimicrobial activity against Gram-negative and Gram-positive bacteria. The authors suggest that the star arrangement affords greater membrane-disrupting capacity than linear forms. For these copolymer SNAPPs, antimicrobial activity increased with respect to the number of star-arms and the star-arm length10. The antimicrobial properties of linear polymers, such as ε- poly(L-lysine) (ε-PLL) have been known for decades and renewed interest in their topical application, for example in wound dressing, is emerging11. Indeed, we have investigated the integration of SNAPP-like star PLL into hydrogels and found potency against S. aureus and E. coli12,13.
In this study, we aimed to investigate the antibacterial and anti-biofilm properties of a series of poly(L-lysine) (PLL)-based polymers in star and linear arrangements, against clinically relevant bacteria, S. aureus (including MRSA) and P. aeruginosa from diabetic foot infections and other wound infections. Importantly, we report highly similar activity of linear and star arrangements of the PLL backbone, which is different to the behaviour reported for linear copolymers of valine and lysine. For PLLs with star-shaped architecture, antimicrobial activity was retained with various hydrophobic amino-acid modifications to the star arms. As such, these peptide-mimetic polymers offer structural flexibility for topical clinical applications and have potential for further development, particularly in the setting of chronic wound infections.
Ethical approval was not sought as it is not required for research on micro-organisms, which are not considered as belonging to the patient or any individual person. No human tissue was used in this study and no additional patient specimen processing or investigations were undertaken. No patient data was collected, processed or stored. Swabs were taken in 2019 at Beaumont Hospital, Dublin, with the patient’s verbal consent, by healthcare staff, from the wound or base of foot ulcers (for DFW) for clinical investigation, as part of their routine clinical care to support the diagnosis of wound infection/diabetic foot infection. Swabs are subsequently routinely processed for recovery and identification of colonizing/infecting organisms by the Beaumont Hospital Microbiology Laboratory. When either S. aureus or P. aeruginosa were identified, pure cultures of these bacteria were provided to the researcher. The study bacteria were received entirely anonymously without any associated patient identifier or data.
This research used the following:
M03079 - H-Lys(Z)-OH - ε-carbobenzyloxy-L-lysine
M02961 - H-Phe-OH - L-phenylalanine
M03002 - H-lle-OH - L-isoleucine
M03100 - H-Tyr(Bzl)-OH - O-benzyl-L-tyrosine
All chemicals and solvents were obtained from Sigma Aldrich (Ireland) unless otherwise noted. ε-carbobenzyloxy-L-lysine (M03079), L-isoleucine (M03002), L-phenylalanine (M02961) and O-benzyl-L-tyrosine (M03100) were purchased from Fluorochem (UK)
The polypeptides and copolypeptides were synthesised via N-carboxyanhydride ring-opening polymerisation (NCA ROP) as previously described14. For synthesis of ε-carbobenzyloxy-L-lysine (ZLL) NCA: ε-carbobenzyloxy-L-lysine (15 g, 53.51 mmol) and α-pinene (147524, 18.22 g, 133.78 mmol) were suspended in 180 mL of dry tetrahydrofuran (THF) and heated under reflux. A solution of triphosgene (330752, 7.15 g, 24.08 mmol) in 30 mL dry THF was added drop-wise to the suspension. The suspension was refluxed until all solids disappeared and the solution became clear (5 hours). The solution was then cooled, filtered and 2/3 of the volume was removed under vacuum. It was then precipitated by addition of 250 mL hexane and stored overnight at -18 °C. The NCA solid was dried, redissolved in dry ethyl acetate and filtered. The NCA solution was then recrystallised thrice in ethyl acetate/hexane (1:1.5) and subsequently washed with hexane to remove any trace impurities. It was vacuum dried to afford a colourless fluffy solid (yield 83%). The NCAs of L-isoleucine (LI), L-phenylalanine (LP) and O-benzyl-L-tyrosine (BLT) were synthesised using the same method. For synthesis of 8arm-star-poly(L-lysine) (G2(8)PLL20) – (representative procedure): ε-carbobenzyloxy-L-lysine (ZLL) NCA (1 g, 2.27 mmol) was dissolved in 15 mL CHCl3 and 3 mL dimethylformamide. G2 poly(propylene imine) (PPI) dendrimer (15.87 mg, 2.04 × 10-2 mmol) was dissolved in 3 mL of CHCl3 and it was then added quickly to the NCA solution. The solution was allowed to stir until Fourier-transform infrared spectroscopy analysis confirmed total consumption of the ZLL NCA monomer. The polymer was then precipitated into excess diethyl ether and dried under vacuum in a dessicator (yield 85%). To deprotect the polypeptide, 500 mg of G2(8)PLL20 was dissolved in 10 mL of trifluoroacetic acid (TFA), followed by dropwise addition of 3 mL of 33 wt% hydrobromic acid (HBr) in acetic acid. The solution was stirred for 18 hours followed by precipitation into excess diethyl ether (150 mL). After drying in vacuo, it was dissolved in deionised water and loaded into a 3500 molecular weight cut off (MWCO) membrane (Thermofisher Scientific, Ireland). Dialysis was performed against deionised water for four days with frequent water replacement, followed by lyophilisation (yield 72%). The other polypepft-irtides were prepared in the same way, using different PPI dendrimers to form G3 and G4 PLL derivatives respectively. The addition of NCA comonomers (LI NCA, LP NCA or BLT NCA) along with ZLL NCA using the same method afforded the statistical copolypeptides containing hydrophobic amino acid repeat units. In this way, three star PLLs were generated: G2(8)PLL20 (8-star-PLL) (G2 = generation 2 poly(propylene imine) dendrimer (PPI) core, (8) = 8 polymeric L-lysine arms with 20 L-lysine subunits per arm, Mn = 21,400 g/mol); G3(16) PLL10 (16-star-PLL) (G3 = generation 3 PPI core, (16) = 16 polymeric L-lysine arms with 10 L-lysine subunits per arm, Mn = 22,300 g/mol); and G4(32)PLL5 (32-star-PLL) (G4 = generation 4 PPI core, (32) = 32 polymeric L-lysine arms with 5 L-lysine subunits per arm, Mn= 24,200 g/mol). The equivalent linear polymer PLL160 was a comparator for this series (no dendrimer core) Mn = 20,600 g/mol. A further G3(16)PLL20 series was generated with an additional five hydrophobic amino acid residues attached to the star arms as follows: G3(16)PLL20-co-PLI5 (G3=generation 3 PPI core, 16 = polymeric L-lysine arms, PLL20 = 20 poly(L-lysine) per arm, co-PLI5 = 5 poly (L-isoleucine) per arm, Mn = 50,300 g/mol, G3(16)PLL20-co-PLP5, co-PLP5 = 5 poly (L-phenylalanine) per arm, Mn = 53,000 g/mol and G3(16)PLL20-co-PLT5, co-PLT5= 5 poly (L-tyrosine) units per arm, Mn = 54,300 g/mol.
A laboratory strain from the American Type Culture Collection, S. aureus ATCC 25923, and the widely used P. aeruginosa strain PAO1 (ATCC 156920) which originates from a wound infection15 were used to establish assay conditions for antimicrobial evaluation of polymers. Two collections of clinical isolates were further evaluated. One consisted of S. aureus and P. aeruginosa recovered from swabs taken as part of routine clinical care from patients with suspected infection during monthly multidisciplinary diabetic foot clinics taking place in Beaumont Hospital, Dublin in 2019 for microbiological diagnosis. The second consisted of 10 S. aureus, five of which were MRSA, which were from confirmed wound infections from the same hospital in the same collection period (2019). All swabs were processed by the Beaumont Hospital Microbiology Laboratory, Dublin and were provided anonymized, as pure cultures. Organism identity was confirmed by matrix-assisted laser desorption/ionization- time-of-flight mass spectrometry (MALDI-TOF) using a MALDI Biotyper (Brüker). MRSA was confirmed by the growth of pink colonies on MRSASelectTM agar (BioRad, UK). In total 30 S. aureus (of which 10 were MRSA) and five P. aeruginosa were collected. For testing novel antimicrobials, clinical isolates were selected randomly from those available to the researchers with numbers chosen as a cross-section, representative of those colonizing and infecting DFIs. The lower numbers of P. aeruginosa available compared to S. aureus recovered are also representative of the trends in recovered pathogens. Within the isolate collection, for determination of statistical significance, e.g. comparison of the activity of linear with star PLLs, biofilm formation, greater than three clinical isolates in each group was considered appropriate.
Assays were performed as described by Forde et al.16 with modifications. Briefly, isolated colonies from overnight growth of S. aureus on Columbia Blood Agar (CBA) or P. aeruginosa on MacConkey agar were suspended in PBS to the density of 0.5 MacFarland Standard using a Densichek Meter (approximately 5 × 107 colony forming units (CFU)/ml). Suspensions were further diluted in the assay buffer (10mM potassium phosphate buffer, pH7.4, containing 0.2% w/v bovine serum albumin (BSA)). Assays contained approximately 105 CFU/ml bacteria and varying concentrations of polymers (0.001μM-10μM) in a total volume of 100μl of assay buffer. Following incubation at 37°C for 1 hour, 900 μl of NaCl (0.95% w/v) was added to assays and 100 μl aliquots were spread onto Mueller Hinton Agar. Plates were incubated at 37°C overnight in a static incubator and resulting colonies were counted. Where numbers were ‘too numerous to count’, further dilutions were plated. Killing activity was determined based on CFU/ml from assays containing PLL polymers (log10 reduction) compared to CFU/ml on control plates containing no polymer. Rifampicin or gentamicin were used as comparators where appropriate against S. aureus and P. aeruginosa, respectively.
Effects of the polymers on biofilms were investigated as described previously17. Colonies from overnight cultures were isolated and used to prepare suspensions to the density of a 0.5 McFarland standard using a Densichek meter (bioMérieux, Ireland). The suspensions were further diluted 1/100 in tryptic soy broth (TSB) containing 0.5% glucose and 100 μL added to 96 well polystyrene plates. Plates were incubated for 24 hours at 37°C. Following washing (3 times with distilled water), the biofilms were treated with various PLL polymers (50 μM) by the addition of 100 μL to wells. Rifampicin or gentamicin (100 μM) was added to biofilm wells as comparators and 100 μl of KPB, pH 7.4 was added to biofilm wells as controls. Wells containing TSB only were included as media controls. The treated plates were incubated at 37°C for 30 minutes. For biofilm viability measurement after treatment, a stock solution of the non-fluorescent redox dye resazurin at 440 μM was diluted 1/5 in TSB and 100 µL was added to each well. Plates were incubated at 37°C for 1 hour in the dark. Fluorescence at 544 nm excitation and 590 nm emission was measured using a Perkin Elmer 2030 Multi-label Reader Viktor X3 spectrophotometer.
Data were analysed and graphs constructed using Microsoft Excel 2016. The log10CFU/ml was determined in controls and treated assays and used to determine the mean log10 reduction in CFU/ml for each PLL structure or antibiotic. For anti-biofilm assays, mean fluorescence intensity units were determined in control and treated assays. Tests of statistical significance of means between groups (control and treated) were by unpaired t-tests using GraphPad Quickcalcs on-line software.
A set of star-shaped poly(L-lysine) structures with 8, 16 and 32 arms were synthesised by the ring-opening polymerisation of ε-carbobenzyloxy-L(lysine) N-carboxyanhydride (ZLL NCA), followed by subsequent deprotection and purification18,19. The total number of PLL units was kept constant at 160 for all star polypeptides resulting in decreasing arm length with increasing number of arms (Figure 1). A linear PLL was also obtained. In addition, 16-arm statistical copolypeptides comprising 20 repeating units of PLL and 5 repeating units of different hydrophobic amino acids were synthesised. The latter were selected from isoleucine, phenylalanine and tyrosine. These and similar structures were previously investigated extensively as delivery tools for applications in tissue engineering where their versatility in delivering therapeutic cargos into mesenchymal stem cells (MSC) has been demonstrated14.
Potent bactericidal activity was demonstrated against the reference S. aureus strain ATCC 25923 and P. aeruginosa strain PAO1. Preliminary testing of 10 μM PLLs, linear (PLL160) Vs star architecture (G2(8)PLL20) for 1 hour against these bacteria resulted in killing greater than the upper limit of the assay (log 5) compared to log 2 reduction in CFU/ml using the antibiotics rifampicin or gentamicin (Figure 2). A lower concentration of 1 μM was therefore investigated to observe differential activity among PLLs. Incubation with PLL-polymers at 1 μM for 1 hour resulted in a 2-4 log reduction in CFU/ml for S. aureus with the greatest activity for the linear PLL160 (4.23 log) and the least for the star architecture with 8 arms around the central core (G2(8)PLL20 = 2.29 log). Improved activity was found as the arm numbers increased to 16 and 32. At 1 μM, 2-3 log reduction in CFU/ml was found for PAO1 with little difference in activity with linear or any of the star architectures, in terms of number of star arms (8, 16, 32) or length of star arms (Figure 3).
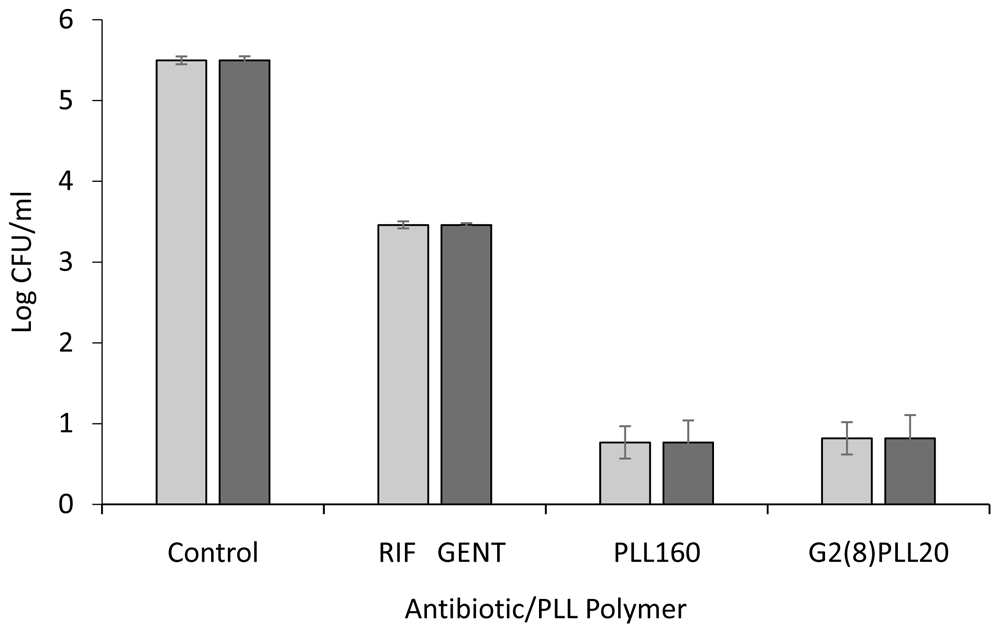
Log CFU/ml for a fixed concentration of bacteria following incubation with 10μM PLL polymers in 10mM potassium phosphate buffer (1 hour, 37oC), 0.2% BSA. S. aureus ATCC 25923 (light grey bars), P. aeruginosa PAO1 (dark grey bars). Rifampicin (RIF) and gentamicin (GENT) were used at 10 μM for comparison. Data shown are the mean ± SEM of three assays carried out in duplicate.
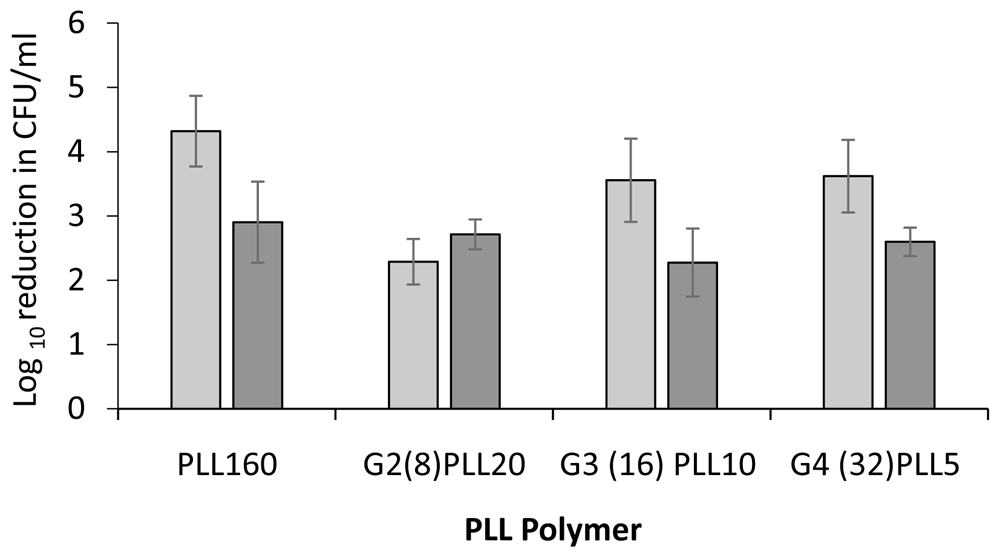
Log reduction in CFU/ml for a fixed concentration of bacteria following incubation with 1μM PLL polymers in 10mM potassium phosphate buffer (1 hour, 37oC), 0.2% BSA. S. aureus ATCC 25923 (light grey bars), P. aeruginosa PAO1 (dark grey bars). Data shown are the mean ± SEM of three assays carried out in duplicate.
Using the G3(16)PLL20 series as a model, further modification of the star arms by the addition of copolypeptides of the amino acids tyrosine, isoleucine or phenylalanine to the poly-lysine arms were made. These modifications did not significantly change the antimicrobial activity against S. aureus (p value ≥ 0.25) which remained at 3-4 log following 1 hour incubation with 1 μM (Figure 4).
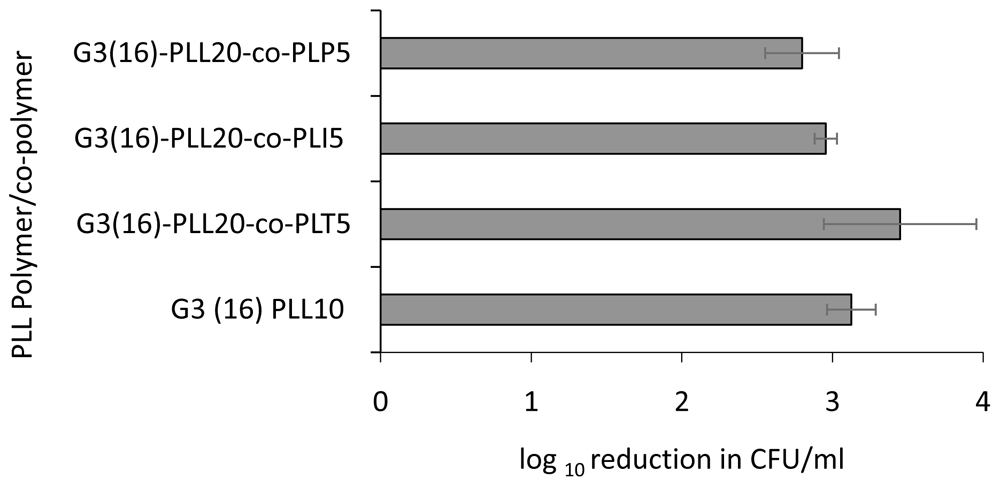
Log reduction in CFU/ml with respect to control for a fixed concentration of S. aureus ATCC 25923, following incubation with 1μM polymer/copolymer in 10mM potassium phosphate buffer (1 hour, 37oC), 0.2% BSA. Data shown are the mean ± SEM of three assays carried out in duplicate.
Clinical isolates recovered from colonized wounds and infections often have different behaviours and properties to laboratory strains. Using PLL160 and G2(8)PLL20 again at 1 μM to reveal potential potency differentials among isolates, both linear and star PLLs remained active against S. aureus (n=10 isolates including 5 MRSA) recovered from confirmed wound infections with mean values of 2.3 and 2.7 log reduction in CFU/ml respectively (Figure 5). As the linear PLL-polymer was the most active at this lower concentration, it was selected for investigation across a further collection of clinical isolates from DFW/DFI. Potent bactericidal activity was observed for S. aureus and P. aeruginosa clinical isolates recovered from DFW/DFI with some variability between isolates (Figure 6). For S. aureus, the mean log reduction was 3.04 ± 0.16 (range 1.86 to 4.22, n=20), for P. aeruginosa isolates the mean log reduction was 3.96 ± 0.82 (range 2.35 to 4.29, n=5).
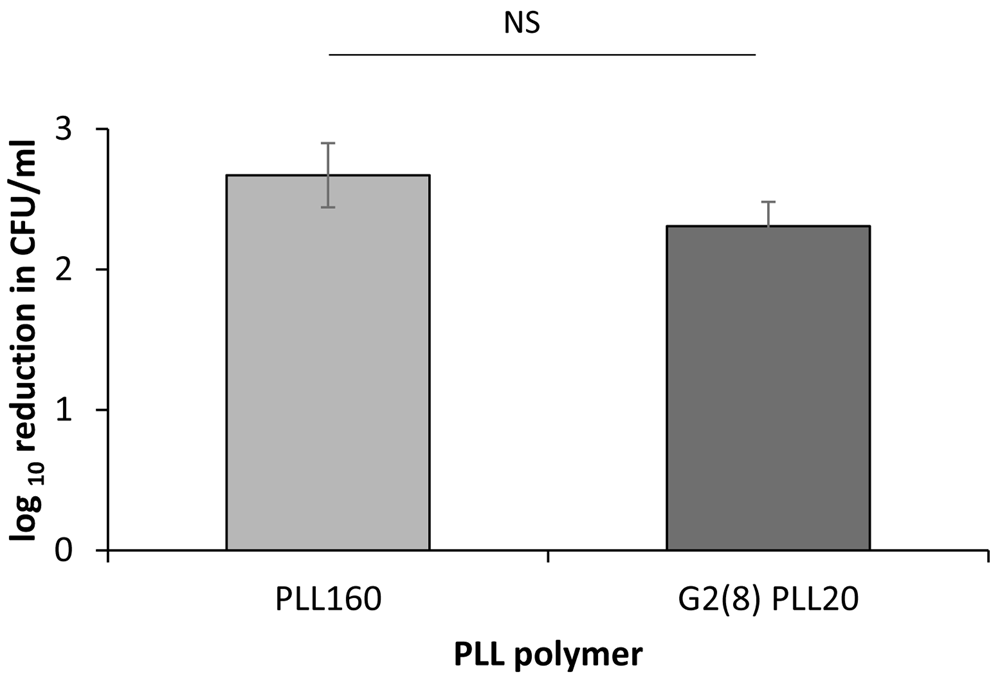
Log reduction in CFU/ml for a fixed concentration of bacteria following incubation with 1μM PLL or G2(8)PLL20 polymer in 10mM potassium phosphate buffer (1 hour, 37oC), 0.2% BSA compared to control (no treatment). Assays carried out in duplicate on three occasions for n=10 S. aureus clinical isolates. NS = not statistically significant, p > 0.1 by unpaired t-test.
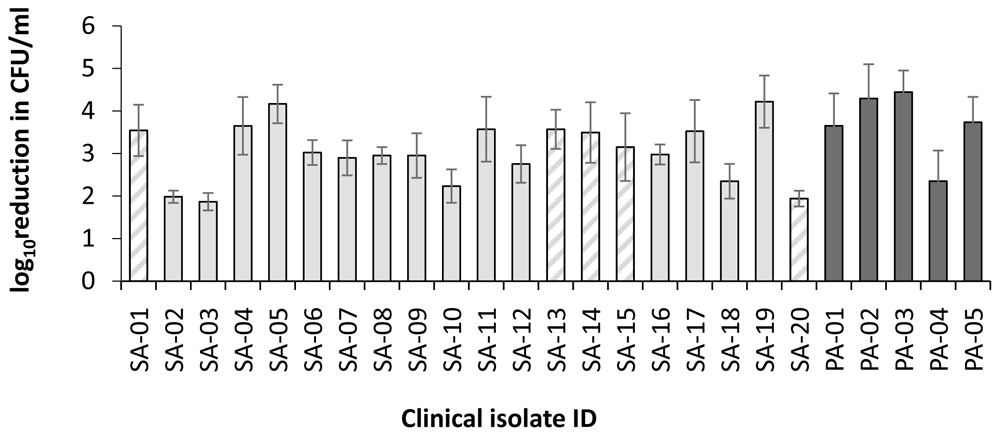
Log reduction in CFU/ml for a fixed concentration of bacteria following incubation with 1 μM PLL160 in 10mM potassium phosphate buffer (1 hour, 37oC), 0.2% BSA compared to control (no treatment). Data shown are the mean ± SEM for assays carried out in duplicate on three occasions per isolate. Hatched bars indicate MRSA isolates.
The G3(16)PLL20 series, including copolymer modifications of the star arms showed statistically significant reduction in biofilm viability over a short time of exposure (30 minutes) to biofilms of S. aureus clinical isolates (p ≤ 0.05 (linear and star G3(16)PLL20 or p ≤ 0.01, co-polymer series). However, only one candidate, G3(16)PLL20-co-PLI5 had anti-biofilm activity against P. aeruginosa isolates (Figure 7). Rifampicin and gentamicin in comparison demonstrated negligible anti-biofilm activity in-vitro under these conditions.
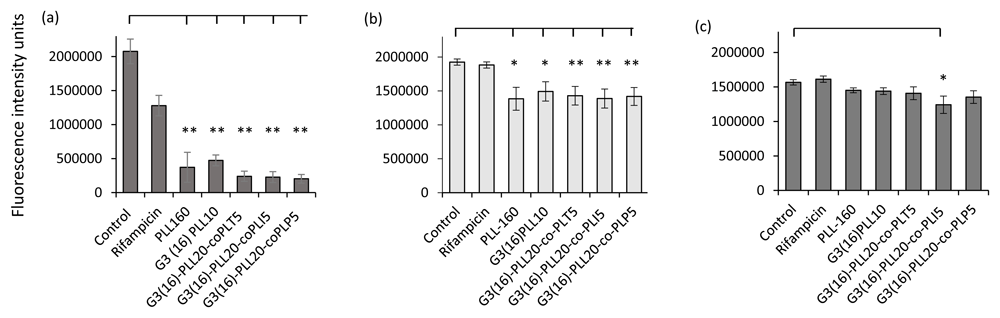
S. aureus reference strain ATCC 25923 (a) and S. aureus DFW/DFI clinical isolates (n=10) (b) or P. aeruginosa DFW/DFI clinical isolates (n=5) recovered from diabetic foot infections. Biofilms were treated with each polymer (50 μM) for 30 minutes at 37oC using at least six replicates per assay. Data shown are the mean ± SEM. Rifampicin/gentamicin concentration = 100 μM, * p ≤ 0.05, ** p ≤ 0.01.
The potential antimicrobial features of star-shaped peptide mimetic polymers has gained much interest since the discovery by Lam et al.9 of low-cost, copolypeptides of lysine and valine synthesised by poly(amido amine) (PAMAM) dendrimer initiated NCA ROP of their corresponding monomers. Those structures have been extensively studied, revealing many features that are critical for their development as alternative antimicrobials, including high potency, low toxicity, activity in in-vivo-like physiological conditions and independent of bacterial antibiotic resistance mechanisms20. Here we report a further family of SNAPPs, Star-PLLs, developed originally as biocompatible polymeric nano-carriers for drug-delivery14,21 applications, as having both antimicrobial and anti-biofilm activity against bacteria recovered from DFW and DFI. Interestingly, in addition, we found the linear equivalent PLL to have similar activity to star-forms.
Diabetic foot infection is the commonest cause of hospitalisation among those with diabetes and the complexity of management may result in lengthened hospital stays. Therefore, to ensure better clinical outcomes for patients, there is a critical need to more effectively manage and treat these infections. A systematic review of antimicrobial agents for chronic wounds, including DFWs, concluded that few systemic agents improved outcomes, but faster healing rates were achieved with additional use of several topical substances4. Synthetic antimicrobial peptides, such as pexiganan, show broad-spectrum activity against DFI pathogens with "all organism" MIC90 of 32 μg/ml reported22. Notably, pexiganan cream used alone was similar in effectiveness to a systemic antibiotic (ofloxacin) in the treatment of mildly infected diabetic foot23. Peptide mimetic polymers with potent bactericidal activity against DFI pathogens, as shown here, may offer enhanced properties for topical wound treatment based on their biocompatible properties which have emerged from studies of their potential in nanocarrier/drug delivery applications.
In the present study, antimicrobial activity was maintained for all PLL polymers investigated with varying architectures from linear to star-shaped, and some with specific copolymer modifications. For all structures, activity was superior to comparator antibiotics, rifampicin or gentamicin, under the same assay conditions. In terms of structure activity relationships (SAR), bactericidal activity was greater when arm number was increased from 8 to 16 but no further increase was noted when further increased to 32. Notably, in this SNAPP series, to maintain equivalence in the number of lysine residues overall, for comparison to the linear polymer, the arm length was decreased (from 20 to 5) with increasing arm number (from 8 to 32). SAR has been studied for a copolymer series of SNAPPs by Shirbin et al. who report antimicrobial activity proportional to the number and length of star arms, albeit with different amino-acid structures. Interestingly, our finding of equivalent antimicrobial activity irrespective of the linear Vs star architecture is markedly different to that reported for star copolymers of L-lysine and L-valine against reference ATCC strains9. Lam et al. reported superior activity of star polymers compared to linear, while, for S. aureus reference strain ATCC25923, we found superior activity of linear PLL160 which was statistically significant (p value ≤ 0.05) compared to the equivalent star 8-mer PLL (G2(8)PLL20). Furthermore, when tested against clinical isolates of S. aureus the bactericidal activity was similar for linear and star PLLs (Figure 5) although inter-isolate variability in activity (PLL160) was evident (Figure 6). As previously highlighted by us, reference strains are not always representative of patient isolates and reliance on reference strains alone may over- or underestimate activity24.
Biofilm production by infecting bacteria is a significant contributor to poorer healing rates and clinical outcomes for patients with wounds infections. All PLL polymers reduced the viability of S. aureus biofilm and this was independent of PLL architectures or modifications. However biofilms of P. aeruginosa were less susceptible to PLLs with only one co-polymer (G3(16)PLL20-co-PLI5) showing statistically significant reduction in metabolic activity. While moderate anti-biofilm activity was found against clinical isolates, the activity against S. aureus reference strain ATCC25923 was significantly greater for all polymers. This finding highlights the need for caution in using reference strains to represent antimicrobial properties against clinical isolates. The structure and physiological properties of biofilms formed by clinical isolates can vary widely under laboratory conditions and even more so when influenced by the in-vivo environment of infection25. The effect of these differences on their response to novel antibiotics and novel antimicrobials should be considered when testing novel anti-biofilm agents.
Our study has limitations. Bactericidal assays were performed in a specific buffer, similar to other published studies and representing physiological conditions (pH 7.4, 10mM potassium phosphate, 37oC). Varying these conditions may alter or reduce activity as has been shown by others20. Further investigation of bactericidal activity under conditions that mimic the pathological environment of wound sites would be important in further development for this clinical application, such as the addition of human serum/wound exudate. Nonetheless, the maintenance of potent activity across clinically relevant isolates, recovered from wounds, demonstrates the antimicrobial potential of this approach. We used an indirect measure of biofilm viability based on metabolic activity rather than viable plate counts and only 24 hour biofilms were tested, showing only moderate to low anti-biofilm activity for this PLL-series against clinically relevant isolates. Improving the anti-biofilm properties of PLL-based and other polymers may be possible iteratively with further structural modifications and should focus on both removal and killing of mature biofilms. As expected based on microbial aetiology, fewer P. aeruginosa clinical isolates were recovered from DFW/DFI compared to S. aureus during the collection period and therefore P. aeruginosa data represent more limited numbers.
Recently Star-PLL, including G4(32)PLL40, have been used to functionalise collagen-based scaffolds with DNA-based therapeutic cargos for bone regeneration/repair applications (bone-morphogenetic protein-2 plasmid (pBMP-2) and vascular endothelial growth factor plasmid (pVEGF)). Osteogenic differentiation of mesenchymal stem cells (MSCs) was demonstrated in-vitro using the PLL-platform and studies in a rat model showed accelerated healing of bone defects in vivo26. The sequelae of diabetic foot wounds and infections may involve abnormal structural defects, e.g. Charcot foot, hammer toe which may further facilitate infection progression27. Adding our finding of bactericidal/anti-biofilm activity relevant specifically to diabetic foot infections, to the properties of these PLL-based biocompatible structures, suggests enhanced and multimodal applications in this setting which will be further explored.
Zenodo: Modified poly(L-lysine)-based structures as novel antimicrobials for diabetic foot infections, an in-vitro study. https://doi.org/10.5281/zenodo.5537160
This project contains the following files:
- Figure 2_PLL_AntimicrobialDFH21V2.xlsx: Bactericidal Activity against S. aureus ATCC25923 and P. aeruginosa PAO1]
- Figure 3_PLL_AntimicrobialDFH21V2.xlsx: Bactericidal activity of poly-l-lysine polymers against S. aureus and P. aeruginosa
- Figure 4_PLL_AntimicrobialDFH21V2.xlsx: Bactericidal activity of poly-L-lysine G3(16) copolymers series with hydrophobic amino acid isoleucine, tyrosine and phenylalanine
- Figure 5_PLL_AntimicrobialDFH21V2.xlsx: Comparison of bactericidal activity of poly-l-lysine polymers, PLL160 and G2(8))PLL20 against S. aureus clinical isolates from wound infections
- Figure 6_PLL_AntimicrobialDFH21V2.xlsx: Bactericidal activity against S. aureus and P. aeruginosa clinical isolates from diabetic foot wounds
- Figure 7_PLL_AntimicrobialDFH21V2.xlsx: Antibiofilm activity against S. aureus ATCC25923, clinical isolates of S. aureus and P. aeruginosa from wounds
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
We thank staff of the Diabetic Foot Clinic, Beaumont Hospital and staff of the Microbiology Laboratory of Beaumont Hospital for assistance with isolate collection.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Microbiome of diabetic foot ulcer infections
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Antimicrobial and antitumor polypeptides
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 12 Jan 22 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Register with HRB Open Research
Already registered? Sign in
Submission to HRB Open Research is open to all HRB grantholders or people working on a HRB-funded/co-funded grant on or since 1 January 2017. Sign up for information about developments, publishing and publications from HRB Open Research.
We'll keep you updated on any major new updates to HRB Open Research
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)