Keywords
COVID-19, coronavirus, immunosuppression, autoimmunity, vasculitis, ANCA, cytokines, immunophenotype
This article is included in the Coronavirus (COVID-19) collection.
COVID-19, coronavirus, immunosuppression, autoimmunity, vasculitis, ANCA, cytokines, immunophenotype
Coronavirus disease 2019 (COVID-19) is a novel, infectious, multi-system disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clinical disease severity is highly variable, ranging from no symptoms, to a mild cough or breathlessness, to severe respiratory failure and death. The COVID-19 pandemic is particularly worrying for people with underlying health conditions, including those taking immunosuppressive medications for autoimmune diseases such as rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), and anti-neutrophil cytoplasmic antibody (ANCA)-associated vasculitis (AAV).
The course of SARS-CoV-2 infection in patients with systemic autoimmune diseases who take immunosuppressive medication is poorly defined. Official guidance on strategies to prevent severe COVID-19 in patients taking immunosuppression is speculative, modelling of disease natural history in this population is ill-informed, and treatment for those who develop COVID-19 relies upon empirically reducing immunosuppression and providing “stress dose” glucocorticoids, without an evidence base to support this approach. Many older adults with autoimmune disease also have co-morbid conditions, including disease-associated organ damage, that might negatively impact their clinical outcomes1,2.
However, early reports indicate that the most damaging facet of COVID-19 is a dysregulated “cytokine storm”3,4, likely driven by dysfunctional neutrophils5–7, suggesting that immunosuppression might confer more benefit than harm. Indeed, previous data suggest that biologic immunosuppressant agents might protect against severe sepsis (OR=0.56)8, and more recently glucocorticoids have shown efficacy as a treatment strategy for COVID-199.
Determining the risk vs. benefit posed by background immunosuppressive therapy, and devising strategies for optimal immunosuppressive management in the setting of COVID-19, has immediate clinical relevance. If immunosuppressive therapy is viewed as a vulnerability factor, rather than as risk-neutral or even protective, then a ward-based ceiling of care might erroneously be applied. Further, immunosuppressive therapy might be discontinued, risking a major disease relapse.
We will address this unmet need and contribute to the national and global COVID-19 response by analysing the impact of immunosuppression on the natural history of, and immune response to, SARS-CoV-2 infection. We will primarily use systemic vasculitis as a prototypical chronic autoimmune disease requiring long-term immunosuppression; however, findings will be broadly revealing about other chronic autoimmune diseases and will inform immunosuppressive medication management during this pandemic. The overarching goal of the DeCOmPRESS study is to determine whether immunosuppressant therapy for chronic autoimmune disease protects against the cytokine storm associated with COVID-19 and reduces the severity of the clinical syndrome.
To achieve this goal, we will examine the following specific aims:
Characterise immune cell subsets altered in COVID infection, comparing immunosuppressed to non-immunosuppressed persons;
Profile serum cytokines in COVID infection, comparing immunosuppressed to non-immunosuppressed persons;
Compare differences in clinical characteristics and outcomes between those who are versus those who are not immunosuppressed at the onset of COVID-19.
The DeCOmPRESS study will use AAV as a chronic autoimmune disease model and take advantage of the existing Irish clinical research infrastructures built around this rare disease. This is a prospective multi-site cohort sub-study of the Rare Kidney Disease (RKD) Biobank, with collection of clinical data and biospecimens as defined in the primary RKD protocol, accessible at https://www.tcd.ie/medicine/thkc/research/rare.php. The RKD Data Management Plan and RKD Registry Data Dictionary are accessible at https://www.tcd.ie/medicine/thkc/decompress/. Our key outcome measure is severe COVID-19 infection, defined as death, ICU admission and/or the need for high-flow (FiO2>30%) oxygen, non-invasive ventilation, or invasive ventilation.
The start date for this project was June 1st, 2020 with an estimated timeframe of 24 months. Existing infrastructure supports the immediate implementation of this work.
The RKD Biobank cohort comprises over 850 members with systemic vasculitis. The three AAV phenotypes are microscopic polyangiitis (MPO), granulomatosis with polyangiitis (GPA), and eosinophilic granulomatosis with polyangiitis (EGPA). This bioresource, which includes about 70% of all known Irish AAV cases, has already supported many published studies. All recruits have provided biological samples during periods of remission and have consented to further sampling and data collection as envisaged for this project. All patients with systemic vasculitis enrolled in the RKD registry who develop probable or confirmed COVID-19 over the next 12–18 months will be included in the DeCOmPRESS study.
Based on modelling data, we anticipate that approximately half of our enrolled patients will develop COVID-19 over the entire duration of the pandemic, and we will capture 75% of these cases (n=306). Based on empiric data (current medication usage in our cohort), we expect that two-thirds (n=202) will meet our definition of exposed (immunosuppressed) and one third (n=104) will not. We expect 52% of cases in the non-immunosuppressed group to develop severe infection. This expected sample size will have 80% power to detect an OR=0.5 (i.e. absolute frequency of severe infection of 35% in immunosuppressed group) or smaller, with 2-tailed alpha of 0.05.
Exposure groups. The patient group for this study comprises RKD Biobank recruits with a diagnosis of systemic vasculitis, as defined by the RKD protocol, and a diagnosis of COVID-19 as confirmed by polymerase chain reaction (PCR) at the time of diagnosis and/or by retrospective serologic testing. Patients with a high clinical suspicion of COVID-19 but no timely PCR test will be included pending confirmatory serologic testing. To estimate the overall period prevalence of SARS-CoV-2 infection in this population, including asymptomatic infection, all RKD recruits will be sampled between September 2020 and March 2021 (i.e., 6 to 12 months after the beginning of the pandemic) to determine the presence of SARS-CoV-2 antibodies. Review of the RKD database will be used to determine type, use, dosage, and frequency of immunosuppressive medications at the time of COVID-19 onset. Immunosuppressive exposure is defined as: current use of >5 mg daily prednisolone or any dose of azathioprine, mycophenolate, avacopan or methotrexate; or rituximab and/or cyclophosphamide use within the previous 6 months. The control group will comprise patients on minimal or no immunosuppression, defined as ≤5 mg daily prednisolone and no other immunosuppressant exposure, as defined above.
Patients with vasculitis previously recruited to the RKD Biobank will be identified, where possible, at the time of COVID-19 diagnosis. Patients will be treated at their local hospital or in the community and clinical data recorded in the RKD database. Blood samples will be obtained for immunophenotyping, serum cytokine measurements, and serologic analysis. This requires an efficient and responsive workflow to allow recruitment at short notice and outside office hours, something that the RKD infrastructure already allows for. However, due to delays in recognising cases or notifying study investigators, some patients might not be recruited until sometime after resolution of COVID-19; only convalescent immunophenotyping will be possible in these cases. A simplified workflow for the DeCOmPRESS study is described in Figure 1.
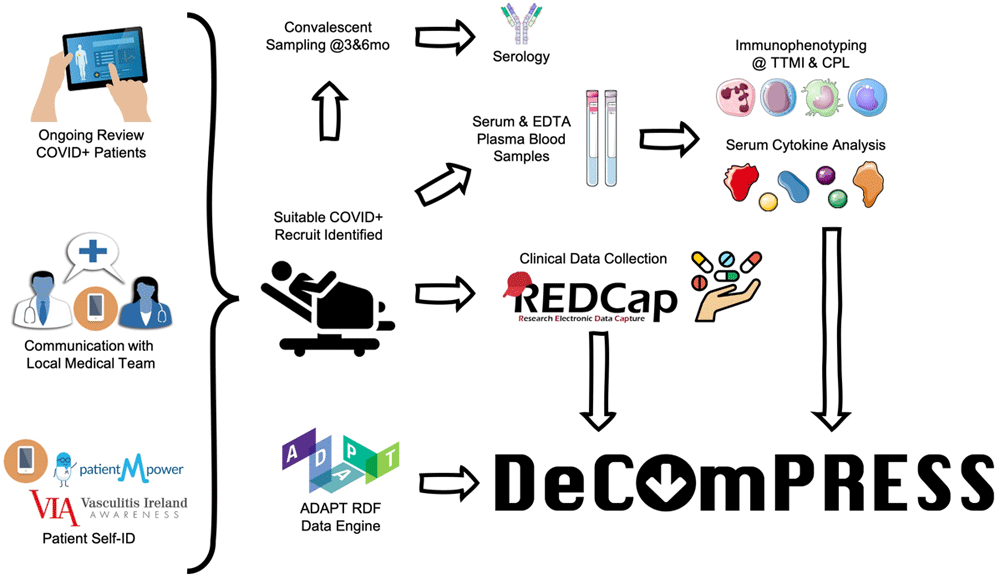
Patients from designated cohorts are recruited at local hospital sites and clinical data recorded in REDCap. Serum and EDTA plasma samples are taken for immunophenotyping and serum cytokine analysis at the Central Pathology Laboratory in St. James's Hospital. Repeat sampling at 3 and 6 months for convalescent antibody testing. Data will be incorporated into the ADAPT RDF data engine for subsequent analysis.
All participants in the DeCOmPRESS study will also be recruited to the RKD Biobank, either previously or at the time of COVID-19 diagnosis. Potential recruits presenting with COVID-19 at participating clinical sites will be reported in a de-identified manner as soon as possible to the DeCOmPRESS study team. There are two primary ways to identify suitable subjects for the DeCOmPRESS study:
1. Contemporaneously upon admission to hospital for COVID-19 treatment.
a. Patients with vasculitis with a confirmed COVID-19 diagnosis (by PCR or serology) will be identified by their local medical team.
b. Those who have not already been recruited to the RKD Biobank will be recruited to this before recruiting to DeCOmPRESS, as per RKD protocol (available at www.tcd.ie/medicine/thkc/research/rare.php).
2. Retrospective screening of local COVID-19 admission logs to identify RKD recruits, and of PCR/serology testing for SARS-CoV-2 in RKD recruits.
Upon identification of an eligible DeCOmPRESS study subject, the study team will follow the steps below:
1. Confirm RKD Main Study ID prior to the collection of clinical data or any study samples.
2. Notify the research team of a potential study subject, without disclosing any identifiable information.
3. Confirm an appropriate time for sample collection and transfer to a central biobank with the research team. This will ideally be less than 4 hours from sample collection to allow for efficient processing.
4. Collect 5 ml serum and 5 ml plasma samples as per RKD protocol.
5. Complete COVID-19 electronic case report form in RKD REDCap database.
Clinical data collection. Clinical data for the DeCOmPRESS study will be captured in an eCRF format using the REDCap platform. REDCap is a secure electronic data capture web platform that stores pseudonymised clinical data for the RKD biobank. The RKD registry database is hosted by Trinity College Dublin (TCD) Research-IT and access permissions are managed by the Principal Investigator (MAL). Comprehensive guidelines for completing the REDCap COVID-19 instrument are available in the RKD Clinical Data Entry standard operating procedure, and video tutorials for REDCap data entry are also available (accessible at www.tcd.ie/medicine/thkc/research/rare.php).
Registration fields are referred to as ‘instruments’ in REDCap. The detailed COVID-19 instrument records information about COVID-19 symptom onset, diagnosis, clinical features, management, and outcomes. Clinical data entry will be completed as soon as possible after patient recruitment to the study and no more than 7 days after biospecimen sampling. Particular attention will be paid to the COVID-19 instrument and prior and current vasculitis encounter medication entries. Patients will be classified as either exposed or unexposed based on immunosuppressant use.
Immunophenotyping and serum cytokine analysis will be carried out at the central Pathology Laboratory (CPL) at St. James’ Hospital Dublin. Serum (minimum 5 ml) and plasma (minimum 5 ml) samples will be taken no more than 24 hours, and ideally within 4 hours, prior to analysis. Immune cell populations (percentages and counts of leukocytes) will be determined by flow cytometry. Serum will be archived to allow subsequent batch measurement of cytokines by ELISA and ELLA Multiplex cytokine assay. Cytokine analysis of historical (obtained during stable vasculitis remission before 1st September 2019), active infection, and convalescent samples will be performed, and compared both within and across participants, to determine the influence of immunosuppression, as well as COVID-19 disease severity, on cytokine levels before, during, and after COVID-19 infection. Additional details of immunophenotyping and serum cytokine analysis procedures are available in the DeCOmPRESS Study Manual (accessible at https://www.tcd.ie/medicine/thkc/decompress/).
Serology. All RKD subjects will be tested for antibodies (IgG, IgM and IgA) to SARS-CoV-2 -spike protein and receptor-binding domain of the spike protein using an in-house developed assay based on work from Mount Sinai10. The samples to be tested will be obtained:
1. 3 and 6 months post suspected or confirmed clinical infection.
2. From all RKD recruits (irrespective of clinical infection) between September 2020 and June 2021.
3. Before September 2019 (from archived serum in the RKD biobank).
A pilot series of patients with PCR-confirmed infection and 20 unselected RKD recruits with samples before and during COVID pandemic were tested for the purpose of this report. We will review patient-reported symptoms suggestive of possible COVID-19, logged via a customised smartphone app within the prior 12 months, and link this to the serological data. This will allow a comprehensive assessment of the impact of immunosuppressive therapy on the antibody response to SARS-CoV-2.
Data analysis will employ the ADAPT Resource Description Framework (RDF) data integration engine and multivariable logistic regression with adjustment for demographics, comorbidities, and vasculitis disease activity11. Information from the multiple DeCOmPRESS data streams integrated into ADAPT’s analysis networks will be stored in a “triplestore” database to facilitate analysis and data sharing. This is a purpose-built database for the storage and retrieval of triples through semantic queries. Semantic triples codify statements about the data in the form of subject–predicate–object expressions. Data from various sources will be formatted appropriately to allow efficient uplift into the triplestore. The primary analysis outcomes will examine differences in immunophenotypes and cytokine profiles, clinical characteristics and outcomes between immunosuppressed and non-immunosuppressed patients. Additional exploratory analyses will include:
1. Differences in patient-reported symptom profiles (immunosuppressed vs. not)
2. Sub-group analyses comparing individual drug regimens e.g. induction vs. maintenance immunosuppression, rituximab vs. cyclophosphamide induction.
3. External comparisons to patients without vasculitis, with or without immunosuppression, who develop COVID-19.
DeCOmPRESS procedures are incorporated into the RKD ethical framework, with prior approval obtained in all RKD recruiting sites. All participants have provided consent for research projects deriving from the samples and clinical data. The DeCOmPRESS sub-project has received additional approval by the RKD Biobank Steering Committee and from the SJH/Tallaght Ethics Committee (REC: 2020-04 List 13 – Amendment (22)). The Health and Safety Authority has directed that university-based non-propagative work using SARS-CoV-2 positive samples should happen at containment level 2 using containment level 3 processes12.
All patients previously enrolled to the RKD Biobank have already provided informed consent for their data and specimens to be used in studies like DeCOmPRESS. Full details are provided in the RKD Biobank protocol. Patient information leaflets are available to download via the following link: https://www.tcd.ie/medicine/thkc/assets/pdf/RKD-Vasculitis-Patient-PIL-ICF-Version-5-07AUG19.pdf. In cases of COVID-19 in a patient with vasculitis who has not already been recruited to the RKD biobank, standard RKD recruitment procedures will be followed.
Descriptive analysis of group characteristics will include number and proportion of cases for categorical variables, mean and standard deviation (SD) for normally distributed continuous variables, and median and inter-quartile range (IQR) for non-normally distributed continuous variables. When examining associations between immunosuppression exposure and COVID-19 outcomes, statistical methods will be employed to account for confounding in vasculitis disease activity, e.g. multivariable logistic regression with handing of disease activity as a stratification variable or interaction term. Additional control groups, within and outside the RKD registry, may be identified and included in the study, pending appropriate ethical approval.
A key objective of DeCOmPRESS is rapid and effective dissemination of research outputs. This has been achieved primarily in conjunction with the Vasculitis Ireland Network (VINE)13, and UK and Ireland Vasculitis Society in conjunction with frequent reviews of state-of-the-art emerging research of relevance to the DeCOmPRESS mission. These are disseminated via a Twitter feed, targeted email distribution lists (managed by the UKIVAS secretariat) and using the VINE website. The DeCOmPRESS study is still recruiting patients and data will be compiled and published 18–24 after project commencement. Initial subject data has contributed to a manuscript with additional UKIVAS patient data and has been submitted for peer review. Full details of the wide-reaching dissemination strategy are outlined in the Data Sharing Plan (accessible at https://www.tcd.ie/medicine/thkc/decompress/).
Current government advisory guidelines for patients taking immunosuppressive therapies encourage increased precautions to protect these vulnerable groups from contracting COVID-19. However, advice regarding how best to manage immunosuppressive therapies, both prior to and after developing COVID-19, are speculative. By comparing the disease course and immune profile of COVID-19 in patients with vasculitis who are versus are not immunosuppressed, the DeCOmPRESS study aims to determine the effects of immunosuppressant therapy on clinical outcomes in this patient group, which in turn will inform patient care. Based on these preliminary results it appears that in-hospital acquisition of COVID-19 during vasculitis investigation and management may be an important means of disease transmission in this population. The prevalence of COVID-19 in the vasculitis patient population appears broadly similar to the wider Irish population.
A key challenge of this study is our recruitment target. Based on epidemic modelling of COVID-19 incidence rates from April 2020, we predicted that approximately half of our cohort would develop COVID-19 over the course of the pandemic, of whom we targeted recruitment of 75% (n=306). However, since the design of this study, vulnerable patients, including those targeted in our study, have been advised to take additional precautions to prevent them from acquiring COVID-19. Further, serial lockdowns, and the emergence of an effective vaccine, might further (fortunately) impact recruitment to this study. Accordingly, in order to answer our research questions, we are exploring adding patients with other rheumatic and dermatological diseases, including those receiving immunosuppressive therapy as defined in this protocol. Further, we are planning more in-depth immunophenotyping to examine in detail the myeloid cell (neutrophil and monocyte) and innate T-cell response to SARS-CoV-2 that could be addressed with smaller patient numbers, should our original target not be reached.
Experience and diversity of study team. The research team were able immediately mobilize the appropriate resources to begin this study on the proposed start date of June 1st, 2020. Our core team and collaborators have a diverse expert knowledge in several areas of clinical and translational research, including immunology, epidemiology, autoimmune disease, and data analysis and integration. A patient representative from Vasculitis Awareness Ireland has also been included in our core research team to guide study design and will ensure that the best interests of patients are considered throughout the study, and that the research questions address issues of relevance to patients.
Access to patient resources via the RKD Biobank. The RKD Biobank comprises over 800 vasculitis patients, including about 70% of all prevalent Irish AAV cases. RKD Biobank recruits have already supported many studies14–17 and provided consent to use their clinical data and biological samples for research. The Vasculitis Ireland Network13 has been recognised by the Department of Health as a centre of expert vasculitis care. Data collection standards are comprehensive and consistent across all clinical study sites.
Powerful data infrastructures. Data generated throughout the DeCOmPRESS study will be uplifted into ADAPT’s RDF data integration engine11. This uses semantic web technology to create a normalised data pool that can be queried and rapidly analysed as a single logical dataset. This will allow rapidly harmonised dissemination of interoperable study data. We have generated a vasculitis-focused COVID-19 dataset, integrated into the RKD database, that adheres to FAIR data principles18. This was developed in collaboration with the FAIRVASC project, which seeks to merge multiple vasculitis registries across Europe, and is interoperable with international datasets. Finally, patients can monitor and report AAV and COVID-19 symptoms through a bespoke smartphone app, developed by Irish SME patientMpower.
No data are associated with this article.
Is the rationale for, and objectives of, the study clearly described?
Yes
Is the study design appropriate for the research question?
Yes
Are sufficient details of the methods provided to allow replication by others?
Yes
Are the datasets clearly presented in a useable and accessible format?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: ANCA-associated vasculitides, biomarker of disease activity and clinical risk stratifications.
Is the rationale for, and objectives of, the study clearly described?
Yes
Is the study design appropriate for the research question?
Yes
Are sufficient details of the methods provided to allow replication by others?
Partly
Are the datasets clearly presented in a useable and accessible format?
Not applicable
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: ANCA vasculitis, lupus nephritis, glomerulonephritis
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 19 Jan 21 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Register with HRB Open Research
Already registered? Sign in
Submission to HRB Open Research is open to all HRB grantholders or people working on a HRB-funded/co-funded grant on or since 1 January 2017. Sign up for information about developments, publishing and publications from HRB Open Research.
We'll keep you updated on any major new updates to HRB Open Research
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)